Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine
- PMID: 20704751
- PMCID: PMC2927562
- DOI: 10.1186/1747-1028-5-19
Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine
Abstract
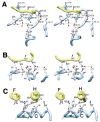
Ubiquitination involves the attachment of ubiquitin to lysine residues on substrate proteins or itself, which can result in protein monoubiquitination or polyubiquitination. Ubiquitin attachment to different lysine residues can generate diverse substrate-ubiquitin structures, targeting proteins to different fates. The mechanisms of lysine selection are not well understood. Ubiquitination by the largest group of E3 ligases, the RING-family E3 s, is catalyzed through co-operation between the non-catalytic ubiquitin-ligase (E3) and the ubiquitin-conjugating enzyme (E2), where the RING E3 binds the substrate and the E2 catalyzes ubiquitin transfer. Previous studies suggest that ubiquitination sites are selected by E3-mediated positioning of the lysine toward the E2 active site. Ultimately, at a catalytic level, ubiquitination of lysine residues within the substrate or ubiquitin occurs by nucleophilic attack of the lysine residue on the thioester bond linking the E2 catalytic cysteine to ubiquitin. One of the best studied RING E3/E2 complexes is the Skp1/Cul1/F box protein complex, SCFCdc4, and its cognate E2, Cdc34, which target the CDK inhibitor Sic1 for K48-linked polyubiquitination, leading to its proteasomal degradation. Our recent studies of this model system demonstrated that residues surrounding Sic1 lysines or lysine 48 in ubiquitin are critical for ubiquitination. This sequence-dependence is linked to evolutionarily conserved key residues in the catalytic region of Cdc34 and can determine if Sic1 is mono- or poly-ubiquitinated. Our studies indicate that amino acid determinants in the Cdc34 catalytic region and their compatibility to those surrounding acceptor lysine residues play important roles in lysine selection. This may represent a general mechanism in directing the mode of ubiquitination in E2 s.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

