Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha
- PMID: 20705240
- PMCID: PMC2921543
- DOI: 10.1016/j.molcel.2010.07.011
Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha
Abstract
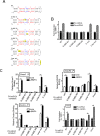
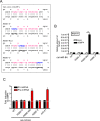
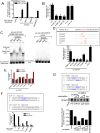
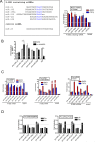
The signal transducers of the transforming growth factor beta (TGFbeta)/bone morphogenetic protein (BMP), the Smads, promote the expression of a subset of miRNAs by facilitating the cleavage reaction by Drosha. The mechanism that limits Smad-mediated processing to a selective group of miRNAs remained hitherto unexplored. In this study, we expand the number of TGFbeta/BMP-regulated miRNAs (T/B-miRs) to 20. Of interest, a majority of T/B-miRs contain a consensus sequence (R-SBE) within the stem region of the primary transcripts of T/B-miRs (pri-T/B-miRs). Here, we demonstrate that Smads directly bind the R-SBE. Mutation of the R-SBE abrogates TGFbeta/BMP-induced recruitment of Smads, Drosha, and DGCR8 to pri-T/B-miRs and impairs their processing, whereas introduction of R-SBE to unregulated pri-miRNAs is sufficient to recruit Smads and to allow regulation by TGFbeta/BMP. Thus, Smads are multifunctional proteins that modulate gene expression transcriptionally through DNA binding and posttranscriptionally through pri-miRNA binding and regulation of miRNA processing.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

Comment in
-
SMADs stimulate miRNA processing.Mol Cell. 2010 Aug 13;39(3):315-6. doi: 10.1016/j.molcel.2010.07.020. Mol Cell. 2010. PMID: 20705233
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

