Nonsense mutations in FAM161A cause RP28-associated recessive retinitis pigmentosa
- PMID: 20705278
- PMCID: PMC2933350
- DOI: 10.1016/j.ajhg.2010.07.018
Nonsense mutations in FAM161A cause RP28-associated recessive retinitis pigmentosa
Abstract
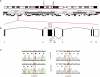
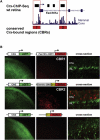
Retinitis pigmentosa (RP) is a degenerative disease of the retina leading to progressive loss of vision and, in many instances, to legal blindness at the end stage. The RP28 locus was assigned in 1999 to the short arm of chromosome 2 by homozygosity mapping in a large Indian family segregating autosomal-recessive RP (arRP). Following a combined approach of chromatin immunoprecipitation and parallel sequencing of genomic DNA, we identified a gene, FAM161A, which was shown to carry a homozygous nonsense mutation (p.Arg229X) in patients from the original RP28 pedigree. Another homozygous FAM161A stop mutation (p.Arg437X) was detected in three subjects from a cohort of 118 apparently unrelated German RP patients. Age at disease onset in these patients was in the second to third decade, with severe visual handicap in the fifth decade and legal blindness in the sixth to seventh decades. FAM161A is a phylogenetically conserved gene, expressed in the retina at relatively high levels and encoding a putative 76 kDa protein of unknown function. In the mouse retina, Fam161a mRNA is developmentally regulated and controlled by the transcription factor Crx, as demonstrated by chromatin immunoprecipitation and organotypic reporter assays on explanted retinas. Fam161a protein localizes to photoreceptor cells during development, and in adult animals it is present in the inner segment as well as the outer plexiform layer of the retina, the synaptic interface between photoreceptors and their efferent neurons. Taken together, our data indicate that null mutations in FAM161A are responsible for the RP28-associated arRP.
2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

