Dosage compensation and demasculinization of X chromosomes in Drosophila
- PMID: 20705467
- PMCID: PMC4511158
- DOI: 10.1016/j.cub.2010.06.076
Dosage compensation and demasculinization of X chromosomes in Drosophila
Abstract
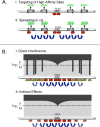
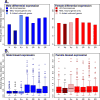
The X chromosome of Drosophila shows a deficiency of genes with male-biased expression, whereas mammalian X chromosomes are enriched for spermatogenesis genes expressed premeiosis and multicopy testis genes. Meiotic X-inactivation and sexual antagonism can only partly account for these patterns. Here, we show that dosage compensation (DC) in Drosophila may contribute substantially to the depletion of male genes on the X. To equalize expression between X-linked and autosomal genes in the two sexes, male Drosophila hypertranscribe their single X, whereas female mammals silence one of their two X chromosomes. We combine fine-scale mapping data of dosage compensated regions with genome-wide expression profiles and show that most male-biased genes on the D. melanogaster X are located outside dosage compensated regions. Additionally, X-linked genes that have newly acquired male-biased expression in D. melanogaster are less likely to be dosage compensated, and parental X-linked genes that gave rise to an autosomal male-biased retrocopy are more likely located within compensated regions. This suggests that DC contributes to the observed demasculinization of X chromosomes in Drosophila, both by limiting the emergence of male-biased expression patterns of existing X genes, and by contributing to gene trafficking of male genes off the X.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures

References
-
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. - PubMed
-
- Khil P, Smirnova N, Romanienko P, Camerini-Otero R. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 2004;36:642–646. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

