ABCG2 transports and transfers heme to albumin through its large extracellular loop
- PMID: 20705604
- PMCID: PMC2963377
- DOI: 10.1074/jbc.M110.139170
ABCG2 transports and transfers heme to albumin through its large extracellular loop
Abstract
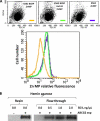
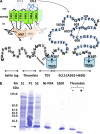
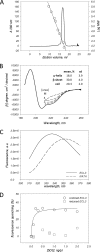
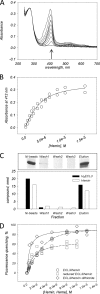
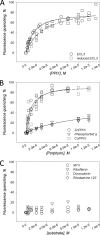
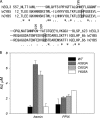
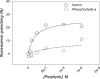
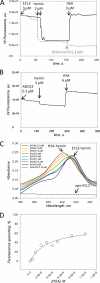
ABCG2 is an ATP-binding cassette (ABC) transporter preferentially expressed by immature human hematopoietic progenitors. Due to its role in drug resistance, its expression has been correlated with a protection role against protoporhyrin IX (PPIX) accumulation in stem cells under hypoxic conditions. We show here that zinc mesoporphyrin, a validated fluorescent heme analog, is transported by ABCG2. We also show that the ABCG2 large extracellular loop ECL3 constitutes a porphyrin-binding domain, which strongly interacts with heme, hemin, PPIX, ZnPPIX, CoPPIX, and much less efficiently with pheophorbide a, but not with vitamin B12. K(d) values are in the range 0.5-3.5 μm, with heme displaying the highest affinity. Nonporphyrin substrates of ABCG2, such as mitoxantrone, doxo/daunorubicin, and riboflavin, do not bind to ECL3. Single-point mutations H583A and C603A inside ECL3 prevent the binding of hemin but hardly affect that of iron-free PPIX. The extracellular location of ECL3 downstream from the transport sites suggests that, after membrane translocation, hemin is transferred to ECL3, which is strategically positioned to release the bound porphyrin to extracellular partners. We show here that human serum albumin could be one of these possible partners as it removes hemin bound to ECL3 and interacts with ABCG2, with a K(d) of about 3 μm.
Figures
References
-
- Dean M., Hamon Y., Chimini G. (2001) J. Lipid Res. 42, 1007–1017 - PubMed
-
- Ross D. D., Yang W., Abruzzo L. V., Dalton W. S., Schneider E., Lage H., Dietel M., Greenberger L., Cole S. P., Doyle L. A. (1999) J. Natl. Cancer Inst. 91, 429–433 - PubMed
-
- Litman T., Brangi M., Hudson E., Fetsch P., Abati A., Ross D. D., Miyake K., Resau J. H., Bates S. E. (2000) J. Cell Sci. 113, 2011–2021 - PubMed
-
- Sarkadi B., Ozvegy-Laczka C., Német K., Váradi A. (2004) FEBS Lett. 567, 116–120 - PubMed
-
- Wang X., Baba M. (2005) Antivir. Chem. Chemother. 16, 213–216 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

