Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex
- PMID: 20705647
- PMCID: PMC3001054
- DOI: 10.1093/nar/gkq668
Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex
Abstract
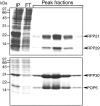
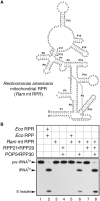
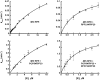
RNase P catalyzes the Mg(2)(+)-dependent 5'-maturation of precursor tRNAs. Biochemical studies on the bacterial holoenzyme, composed of one catalytic RNase P RNA (RPR) and one RNase P protein (RPP), have helped understand the pleiotropic roles (including substrate/Mg(2+) binding) by which a protein could facilitate RNA catalysis. As a model for uncovering the functional coordination among multiple proteins that aid an RNA catalyst, we use archaeal RNase P, which comprises one catalytic RPR and at least four RPPs. Exploiting our previous finding that these archaeal RPPs function as two binary RPP complexes (POP5•RPP30 and RPP21•RPP29), we prepared recombinant RPP pairs from three archaea and established interchangeability of subunits through homologous/heterologous assemblies. Our finding that archaeal POP5•RPP30 reconstituted with bacterial and organellar RPRs suggests functional overlap of this binary complex with the bacterial RPP and highlights their shared recognition of a phylogenetically-conserved RPR catalytic core, whose minimal attributes we further defined through deletion mutagenesis. Moreover, single-turnover kinetic studies revealed that while POP5•RPP30 is solely responsible for enhancing the RPR's rate of precursor tRNA cleavage (by 60-fold), RPP21•RPP29 contributes to increased substrate affinity (by 16-fold). Collectively, these studies provide new perspectives on the functioning and evolution of an ancient, catalytic ribonucleoprotein.
Figures
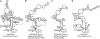

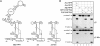

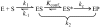
References
-
- Liu F, Altman S. Ribonuclease P. Protein Reviews Series. Springer-Verlag: New York; 2010.
-
- Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. - PubMed
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

