Mapping of the nuclear matrix-bound chromatin hubs by a new M3C experimental procedure
- PMID: 20705651
- PMCID: PMC3001081
- DOI: 10.1093/nar/gkq712
Mapping of the nuclear matrix-bound chromatin hubs by a new M3C experimental procedure
Abstract
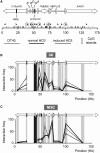
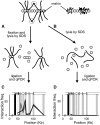
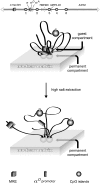
We have developed an experimental procedure to analyze the spatial proximity of nuclear matrix-bound DNA fragments. This protocol, referred to as Matrix 3C (M3C), includes a high salt extraction of nuclei, the removal of distal parts of unfolded DNA loops using restriction enzyme treatment, ligation of the nuclear matrix-bound DNA fragments and a subsequent analysis of ligation frequencies. Using the M3C procedure, we have demonstrated that CpG islands of at least three housekeeping genes that surround the chicken α-globin gene domain are assembled into a complex (presumably, a transcription factory) that is stabilized by the nuclear matrix in both erythroid and non-erythroid cells. In erythroid cells, the regulatory elements of the α-globin genes are attracted to this complex to form a new assembly: an active chromatin hub that is linked to the pre-existing transcription factory. The erythroid-specific part of the assembly is removed by high salt extraction. Based on these observations, we propose that mixed transcription factories that mediate the transcription of both housekeeping and tissue-specific genes are composed of a permanent compartment containing integrated into the nuclear matrix promoters of housekeeping genes and a 'guest' compartment where promoters and regulatory elements of tissue-specific genes can be temporarily recruited.
Figures


Similar articles
-
Spatial configuration of the chicken alpha-globin gene domain: immature and active chromatin hubs.Nucleic Acids Res. 2008 Aug;36(14):4629-40. doi: 10.1093/nar/gkn429. Epub 2008 Jul 11. Nucleic Acids Res. 2008. PMID: 18621783 Free PMC article.
-
The 33 kb transcript of the chicken alpha-globin gene domain is part of the nuclear matrix.J Cell Biochem. 2004 Jun 1;92(3):445-57. doi: 10.1002/jcb.20066. J Cell Biochem. 2004. PMID: 15156557
-
Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes.Mol Cell Biol. 2006 Jul;26(13):5096-105. doi: 10.1128/MCB.02454-05. Mol Cell Biol. 2006. PMID: 16782894 Free PMC article.
-
Close encounters of the 3C kind: long-range chromatin interactions and transcriptional regulation.Brief Funct Genomic Proteomic. 2009 Jul;8(4):297-309. doi: 10.1093/bfgp/elp016. Epub 2009 Jun 17. Brief Funct Genomic Proteomic. 2009. PMID: 19535505 Review.
-
Chromatin domains and regulation of gene expression: familiar and enigmatic clusters of chicken globin genes.Crit Rev Eukaryot Gene Expr. 2001;11(1-3):227-42. Crit Rev Eukaryot Gene Expr. 2001. PMID: 11693962 Review.
Cited by
-
Unmethylated CpG islands are clustered inside the interphase human cell nuclei.Dokl Biochem Biophys. 2012 Mar-Apr;443:123-6. doi: 10.1134/S1607672912020160. Epub 2012 May 5. Dokl Biochem Biophys. 2012. PMID: 22562640 No abstract available.
-
Transcription factories in the context of the nuclear and genome organization.Nucleic Acids Res. 2011 Nov;39(21):9085-92. doi: 10.1093/nar/gkr683. Epub 2011 Aug 31. Nucleic Acids Res. 2011. PMID: 21880598 Free PMC article. Review.
-
Nuclear scaffold attachment sites within ENCODE regions associate with actively transcribed genes.PLoS One. 2011 Mar 14;6(3):e17912. doi: 10.1371/journal.pone.0017912. PLoS One. 2011. PMID: 21423757 Free PMC article.
-
Transcription factories.Front Genet. 2012 Oct 23;3:221. doi: 10.3389/fgene.2012.00221. eCollection 2012. Front Genet. 2012. PMID: 23109938 Free PMC article.
-
Large-scale organization of ribosomal DNA chromatin is regulated by Tip5.Nucleic Acids Res. 2013 May 1;41(10):5251-62. doi: 10.1093/nar/gkt218. Epub 2013 Apr 10. Nucleic Acids Res. 2013. PMID: 23580549 Free PMC article.
References
-
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. - PubMed
-
- Splinter E, Grosveld F, de Laat W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. - PubMed
-
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. - PubMed
-
- de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. - PubMed
-
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F. Three-dimensional organization of gene expression in erythroid cells. Curr. Top. Dev. Biol. 2008;82:117–139. - PubMed

