SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer
- PMID: 20708153
- PMCID: PMC3954541
- DOI: 10.1016/j.ccr.2010.06.018
SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer
Abstract
Regulators of mitosis have been successfully targeted to enhance response to taxane chemotherapy. Here, we show that the salt inducible kinase 2 (SIK2) localizes at the centrosome, plays a key role in the initiation of mitosis, and regulates the localization of the centrosome linker protein, C-Nap1, through S2392 phosphorylation. Interference with the known SIK2 inhibitor PKA induced SIK2-dependent centrosome splitting in interphase while SIK2 depletion blocked centrosome separation in mitosis, sensitizing ovarian cancers to paclitaxel in culture and in xenografts. Depletion of SIK2 also delayed G1/S transition and reduced AKT phosphorylation. Higher expression of SIK2 significantly correlated with poor survival in patients with high-grade serous ovarian cancers. We believe these data identify SIK2 as a plausible target for therapy in ovarian cancers.
2010 Elsevier Inc. All rights reserved.
Figures





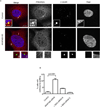

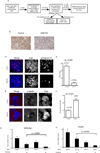
Comment in
-
A centrosome kinase modulates antitumor drug sensitivity.Cancer Cell. 2010 Aug 9;18(2):99-100. doi: 10.1016/j.ccr.2010.07.008. Cancer Cell. 2010. PMID: 20708149 Free PMC article.
References
-
- Bekier ME, Fischbach R, Lee J, Taylor WR. Length of mitotic arrest induced by microtubule-stabilizing drugs determines cell death after mitotic exit. Mol Cancer Ther. 2009;8:1646–1654. - PubMed
-
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. - PubMed
-
- Carlson CR, Witczak O, Vossebein L, Labbe JC, Skalhegg BS, Keryer G, Herberg FW, Collas P, Tasken K. CDK1-mediated phosphorylation of the RIIalpha regulatory subunit of PKA works as a molecular switch that promotes dissociation of RIIalpha from centrosomes at mitosis. J Cell Sci. 2001;114:3243–3254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

