Maintenance of duplicate genes and their functional redundancy by reduced expression
- PMID: 20708291
- PMCID: PMC2942974
- DOI: 10.1016/j.tig.2010.07.002
Maintenance of duplicate genes and their functional redundancy by reduced expression
Abstract
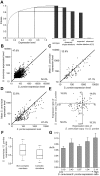
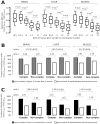
Although evolutionary theories predict functional divergence between duplicate genes, many old duplicates still maintain a high degree of functional similarity and are synthetically lethal or sick, an observation that has puzzled many geneticists. We propose that expression reduction, a special type of subfunctionalization, facilitates the retention of duplicates and the conservation of their ancestral functions. Consistent with this hypothesis, gene expression data from both yeasts and mammals show a substantial decrease in the level of gene expression after duplication. Whereas the majority of the expression reductions are likely to be neutral, some are apparently beneficial to rebalancing gene dosage after duplication.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Ohno S. Evolution by Gene Duplication. Springer; Verlag: 1970.
-
- Zhang J. Evolution by gene duplication-an update. Trends Ecol. Evol. 2003;18:292–298.
-
- DeLuna A, et al. Exposing the fitness contribution of duplicated genes. Nat Genet. 2008;40:676–681. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

