The 3D structures of VDAC represent a native conformation
- PMID: 20708406
- PMCID: PMC2933295
- DOI: 10.1016/j.tibs.2010.03.005
The 3D structures of VDAC represent a native conformation
Abstract
The most abundant protein of the mitochondrial outer membrane is the voltage-dependent anion channel (VDAC), which facilitates the exchange of ions and molecules between mitochondria and cytosol and is regulated by interactions with other proteins and small molecules. VDAC has been studied extensively for more than three decades, and last year three independent investigations revealed a structure of VDAC-1 exhibiting 19 transmembrane beta-strands, constituting a unique structural class of beta-barrel membrane proteins. Here, we provide a historical perspective on VDAC research and give an overview of the experimental design used to obtain these structures. Furthermore, we validate the protein refolding approach and summarize the biochemical and biophysical evidence that links the 19-stranded structure to the native form of VDAC.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
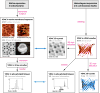

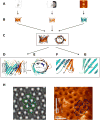
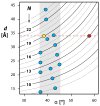
References
-
- Parsons DF, et al. Characteristics of isolated and purified preparations of outer and inner membranes of mitochondria. Ann NY Acad Sci. 1966;137:643–666. - PubMed
-
- Schein SJ, et al. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from Paramecium mitochondria. J Membr Biol. 1976;30:99–120. - PubMed
-
- Mannella CA, Colombini M. Evidence that the crystalline arrays in the outer membrane of Neurospora mitochondria are composed of the voltage-dependent channel protein. Biochim Biophys Acta. 1984;774:206–214. - PubMed
-
- Guo XW, et al. Molecular design of the voltage-dependent, anion-selective channel in the mitochondrial outer membrane. J Struct Biol. 1995;114:41–59. - PubMed
-
- Colombini M. A candidate for the permeability pathway of the outer mitochondrial membrane. Nature. 1979;279:643–645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

