beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2
- PMID: 20708585
- PMCID: PMC2923644
- DOI: 10.1016/j.devcel.2010.07.007
beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2
Abstract
An emerging concept in development is that transcriptional poising presets patterns of gene expression in a manner that reflects a cell's developmental potential. However, it is not known how certain loci are specified in the embryo to establish poised chromatin architecture as the developmental program unfolds. We find that, in the context of transcriptional quiescence prior to the midblastula transition in Xenopus, dorsal specification by the Wnt/beta-catenin pathway is temporally uncoupled from the onset of dorsal target gene expression, and that beta-catenin establishes poised chromatin architecture at target promoters. beta-catenin recruits the arginine methyltransferase Prmt2 to target promoters, thereby establishing asymmetrically dimethylated H3 arginine 8 (R8). Recruitment of Prmt2 to beta-catenin target genes is necessary and sufficient to establish the dorsal developmental program, indicating that Prmt2-mediated histone H3(R8) methylation plays a critical role downstream of beta-catenin in establishing poised chromatin architecture and marking key organizer genes for later expression.
2010 Elsevier Inc. All rights reserved.
Figures
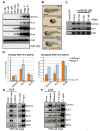
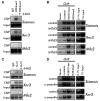
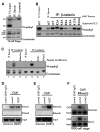
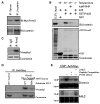
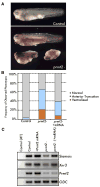
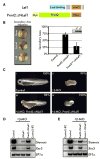
Comment in
-
On the fast track to organizer gene expression.Dev Cell. 2010 Aug 17;19(2):190-2. doi: 10.1016/j.devcel.2010.07.020. Dev Cell. 2010. PMID: 20708581 Free PMC article.
-
Development: Patterning factor poises genes for expression.Nat Rev Genet. 2010 Oct;11(10):668. doi: 10.1038/nrg2869. Epub 2010 Sep 14. Nat Rev Genet. 2010. PMID: 20838410 No abstract available.
References
-
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II Accumulates at Promoters of Growth Genes During Developmental Arrest. Science. 2009;324:92–94. - PubMed
-
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
-
- Belenkaya TY, Han C, Standley HJ, Lin X, Houston DW, Heasman J, Lin X. pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Development. 2002;129:4089–4101. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

