CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule
- PMID: 20708587
- PMCID: PMC3156696
- DOI: 10.1016/j.devcel.2010.07.016
CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule
Abstract
Spatial regulation of microtubule (MT) dynamics contributes to cell polarity and cell division. MT rescue, in which a MT stops shrinking and reinitiates growth, is the least understood aspect of MT dynamics. Cytoplasmic Linker Associated Proteins (CLASPs) are a conserved class of MT-associated proteins that contribute to MT stabilization and rescue in vivo. We show here that the Schizosaccharomyces pombe CLASP, Cls1p, is a homodimer that binds an alphabeta-tubulin heterodimer through conserved TOG-like domains. In vitro, CLASP increases MT rescue frequency, decreases MT catastrophe frequency, and moderately decreases MT disassembly rate. CLASP binds stably to the MT lattice, recruits tubulin, and locally promotes rescues. Mutations in the CLASP TOG domains demonstrate that tubulin binding is critical for its rescue activity. We propose a mechanism for rescue in which CLASP-tubulin dimer complexes bind along the MT lattice and reverse MT depolymerization with their bound tubulin dimer.
2010 Elsevier Inc. All rights reserved.
Figures



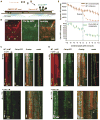
I. 6 μM tubulin: dynamic MTs show slow assembly, rapid disassembly, and frequent catastrophes.
II. 50 nM XMAP215 + 6 μM tubulin: dynamic MTs show rapid MT assembly and frequent catastrophes.
III-V. 40 nM Cls1p + 6 μM tubulin: Panel III shows a relatively slow assembly MT rate and no catastrophes. Panel IV shows a dynamic MT assembling slowly with two catastrophes, each followed by a rescue. Panel V shows a dynamic MT assembling slowly and a catastrophe that leads to extended disassembly, which is followed by a rescue.



References
-
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. - PubMed
-
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. Epub 2008 Mar 2005. - PubMed
-
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

