Modeling studies of chromatin fiber structure as a function of DNA linker length
- PMID: 20709077
- PMCID: PMC2966533
- DOI: 10.1016/j.jmb.2010.07.057
Modeling studies of chromatin fiber structure as a function of DNA linker length
Abstract
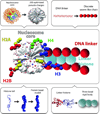
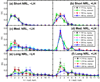
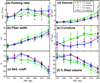
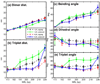
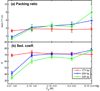
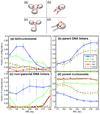
Chromatin fibers encountered in various species and tissues are characterized by different nucleosome repeat lengths (NRLs) of the linker DNA connecting the nucleosomes. While single cellular organisms and rapidly growing cells with high protein production have short NRL ranging from 160 to 189 bp, mature cells usually have longer NRLs ranging between 190 and 220 bp. Recently, various experimental studies have examined the effect of NRL on the internal organization of chromatin fiber. Here, we investigate by mesoscale modeling of oligonucleosomes the folding patterns for different NRL, with and without linker histone (LH), under typical monovalent salt conditions using both one-start solenoid and two-start zigzag starting configurations. We find that short to medium NRL chromatin fibers (173 to 209 bp) with LH condense into zigzag structures and that solenoid-like features are viable only for longer NRLs (226 bp). We suggest that medium NRLs are more advantageous for packing and various levels of chromatin compaction throughout the cell cycle than their shortest and longest brethren; the former (short NRLs) fold into narrow fibers, while the latter (long NRLs) arrays do not easily lead to high packing ratios due to possible linker DNA bending. Moreover, we show that the LH has a small effect on the condensation of short-NRL arrays but has an important condensation effect on medium-NRL arrays, which have linker lengths similar to the LH lengths. Finally, we suggest that the medium-NRL species, with densely packed fiber arrangements, may be advantageous for epigenetic control because their histone tail modifications can have a greater effect compared to other fibers due to their more extensive nucleosome interaction network.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Chromatin fiber polymorphism triggered by variations of DNA linker lengths.Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):8061-6. doi: 10.1073/pnas.1315872111. Epub 2014 May 20. Proc Natl Acad Sci U S A. 2014. PMID: 24847063 Free PMC article.
-
Mesoscale simulations of two nucleosome-repeat length oligonucleosomes.Phys Chem Chem Phys. 2009 Dec 7;11(45):10729-37. doi: 10.1039/b918629h. Epub 2009 Oct 20. Phys Chem Chem Phys. 2009. PMID: 20145817 Free PMC article.
-
Sensitive effect of linker histone binding mode and subtype on chromatin condensation.Nucleic Acids Res. 2019 Jun 4;47(10):4948-4957. doi: 10.1093/nar/gkz234. Nucleic Acids Res. 2019. PMID: 30968131 Free PMC article.
-
Structural insights of nucleosome and the 30-nm chromatin fiber.Curr Opin Struct Biol. 2016 Feb;36:106-15. doi: 10.1016/j.sbi.2016.01.013. Epub 2016 Feb 9. Curr Opin Struct Biol. 2016. PMID: 26872330 Review.
-
Linking Chromatin Fibers to Gene Folding by Hierarchical Looping.Biophys J. 2017 Feb 7;112(3):434-445. doi: 10.1016/j.bpj.2017.01.003. Epub 2017 Jan 31. Biophys J. 2017. PMID: 28153411 Free PMC article. Review.
Cited by
-
Topological polymorphism of the two-start chromatin fiber.Biophys J. 2015 May 19;108(10):2591-2600. doi: 10.1016/j.bpj.2015.04.015. Biophys J. 2015. PMID: 25992737 Free PMC article.
-
Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub.Proc Natl Acad Sci U S A. 2019 Mar 12;116(11):4955-4962. doi: 10.1073/pnas.1816424116. Epub 2019 Feb 4. Proc Natl Acad Sci U S A. 2019. PMID: 30718394 Free PMC article.
-
In diverse conditions, intrinsic chromatin condensates have liquid-like material properties.Proc Natl Acad Sci U S A. 2023 May 2;120(18):e2218085120. doi: 10.1073/pnas.2218085120. Epub 2023 Apr 24. Proc Natl Acad Sci U S A. 2023. PMID: 37094140 Free PMC article.
-
In silico nanoscope to study the interplay of genome organization and transcription regulation.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf189. doi: 10.1093/nar/gkaf189. Nucleic Acids Res. 2025. PMID: 40114377 Free PMC article.
-
Insights into chromatin fibre structure by in vitro and in silico single-molecule stretching experiments.Biochem Soc Trans. 2013 Apr;41(2):494-500. doi: 10.1042/BST20120349. Biochem Soc Trans. 2013. PMID: 23514142 Free PMC article. Review.
References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
-
- Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. - PubMed
-
- Bates DL, Jonathan P, Butler G, Pearson EC, Thomas JO. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur. J. Biochem. 1981;119:469–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

