Functional characterization of pGKT2, a 182-kilobase plasmid containing the xplAB genes, which are involved in the degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine by Gordonia sp. strain KTR9
- PMID: 20709853
- PMCID: PMC2950474
- DOI: 10.1128/AEM.01217-10
Functional characterization of pGKT2, a 182-kilobase plasmid containing the xplAB genes, which are involved in the degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine by Gordonia sp. strain KTR9
Abstract
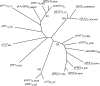
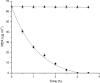
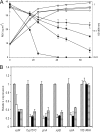
Several microorganisms have been isolated that can transform hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), a cyclic nitramine explosive. To better characterize the microbial genes that facilitate this transformation, we sequenced and annotated a 182-kb plasmid, pGKT2, from the RDX-degrading strain Gordonia sp. KTR9. This plasmid carries xplA, encoding a protein sharing up to 99% amino acid sequence identity with characterized RDX-degrading cytochromes P450. Other genes that cluster with xplA are predicted to encode a glutamine synthase-XplB fusion protein, a second cytochrome P450, Cyp151C, and XplR, a GntR-type regulator. Rhodococcus jostii RHA1 expressing xplA from KTR9 degraded RDX but did not utilize RDX as a nitrogen source. Moreover, an Escherichia coli strain producing XplA degraded RDX but a strain producing Cyp151C did not. KTR9 strains cured of pGKT2 did not transform RDX. Physiological studies examining the effects of exogenous nitrogen sources on RDX degradation in strain KTR9 revealed that ammonium, nitrite, and nitrate each inhibited RDX degradation by up to 79%. Quantitative real-time PCR analysis of glnA-xplB, xplA, and xplR showed that transcript levels were 3.7-fold higher during growth on RDX than during growth on ammonium and that this upregulation was repressed in the presence of various inorganic nitrogen sources. Overall, the results indicate that RDX degradation by KTR9 is integrated with central nitrogen metabolism and that the uptake of RDX by bacterial cells does not require a dedicated transporter.
Figures


Similar articles
-
Investigating differences in the ability of XplA/B-containing bacteria to degrade the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX).FEMS Microbiol Lett. 2017 Aug 1;364(14). doi: 10.1093/femsle/fnx144. FEMS Microbiol Lett. 2017. PMID: 28854671
-
The essential role of nitrogen limitation in expression of xplA and degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) in Gordonia sp. strain KTR9.Appl Microbiol Biotechnol. 2015 Jan;99(1):459-67. doi: 10.1007/s00253-014-6013-z. Epub 2014 Aug 22. Appl Microbiol Biotechnol. 2015. PMID: 25142696
-
Horizontal gene transfer (HGT) as a mechanism of disseminating RDX-degrading activity among Actinomycete bacteria.J Appl Microbiol. 2011 Jun;110(6):1449-59. doi: 10.1111/j.1365-2672.2011.04995.x. Epub 2011 Mar 30. J Appl Microbiol. 2011. PMID: 21395947
-
The explosive-degrading cytochrome P450 XplA: biochemistry, structural features and prospects for bioremediation.Biochim Biophys Acta. 2011 Jan;1814(1):230-6. doi: 10.1016/j.bbapap.2010.07.004. Epub 2010 Jul 17. Biochim Biophys Acta. 2011. PMID: 20624490 Review.
-
Microbial degradation and toxicity of hexahydro-1,3,5-trinitro-1,3,5-triazine.J Microbiol Biotechnol. 2012 Oct;22(10):1311-23. doi: 10.4014/jmb.1203.04002. J Microbiol Biotechnol. 2012. PMID: 23075780 Review.
Cited by
-
Analysis of the xplAB-containing gene cluster involved in the bacterial degradation of the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine.Appl Environ Microbiol. 2014 Nov;80(21):6601-10. doi: 10.1128/AEM.01818-14. Epub 2014 Aug 15. Appl Environ Microbiol. 2014. PMID: 25128343 Free PMC article.
-
Biodegradation of insensitive munition formulations IMX101 and IMX104 in surface soils.J Ind Microbiol Biotechnol. 2017 Jul;44(7):987-995. doi: 10.1007/s10295-017-1930-3. Epub 2017 Mar 3. J Ind Microbiol Biotechnol. 2017. PMID: 28258407
-
Metagenomic insights into the RDX-degrading potential of the ovine rumen microbiome.PLoS One. 2014 Nov 10;9(11):e110505. doi: 10.1371/journal.pone.0110505. eCollection 2014. PLoS One. 2014. PMID: 25383623 Free PMC article.
-
Stable isotope probing reveals the importance of Comamonas and Pseudomonadaceae in RDX degradation in samples from a Navy detonation site.Environ Sci Pollut Res Int. 2015 Jul;22(13):10340-50. doi: 10.1007/s11356-015-4256-6. Epub 2015 Feb 28. Environ Sci Pollut Res Int. 2015. PMID: 25721530
-
Crucial role of a dicarboxylic motif in the catalytic center of yeast RNA polymerases.Curr Genet. 2011 Oct;57(5):327-34. doi: 10.1007/s00294-011-0350-6. Epub 2011 Jul 15. Curr Genet. 2011. PMID: 21761155
References
-
- Adrian, N. R., and C. M. Arnett. 2004. Anaerobic biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Acetobacterium malicum strain HAAP-1 isolated from a methanogenic mixed culture. Curr. Microbiol. 48:332-340. - PubMed
-
- Adrian, N. R., C. M. Arnett, and R. F. Hickey. 2003. Stimulating the anaerobic biodegradation of explosives by the addition of hydrogen or electron donors that produce hydrogen. Water Res. 37:3499-3507. - PubMed
-
- Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

