Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence
- PMID: 20709895
- PMCID: PMC2950490
- DOI: 10.1128/JB.00883-10
Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence
Abstract
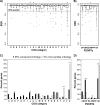
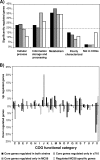
Neisseria meningitidis serogroup B strains are responsible for most meningococcal cases in the industrialized countries, and strains belonging to the clonal complex ST-41/44 are among the most prevalent serogroup B strains in carriage and disease. Here, we report the first genome and transcriptome comparison of a serogroup B carriage strain from the clonal complex ST-41/44 to the serogroup B disease strain MC58 from the clonal complex ST-32. Both genomes are highly colinear, with only three major genome rearrangements that are associated with the integration of mobile genetic elements. They further differ in about 10% of their gene content, with the highest variability in gene presence as well as gene sequence found for proteins involved in host cell interactions, including Opc, NadA, TonB-dependent receptors, RTX toxin, and two-partner secretion system proteins. Whereas housekeeping genes coding for metabolic functions were highly conserved, there were considerable differences in their expression pattern upon adhesion to human nasopharyngeal cells between both strains, including differences in energy metabolism and stress response. In line with these genomic and transcriptomic differences, both strains also showed marked differences in their in vitro infectivity and in serum resistance. Taken together, these data support the concept of a polygenic nature of meningococcal virulence comprising differences in the repertoire of adhesins as well as in the regulation of metabolic genes and suggest a prominent role for immune selection and genetic drift in shaping the meningococcal genome.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Arber, W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24:1-7. - PubMed
-
- Bentley, S. D., G. S. Vernikos, L. A. Snyder, C. Churcher, C. Arrowsmith, T. Chillingworth, A. Cronin, P. H. Davis, N. E. Holroyd, K. Jagels, M. Maddison, S. Moule, E. Rabbinowitsch, S. Sharp, L. Unwin, S. Whitehead, M. A. Quail, M. Achtman, B. Barrell, N. J. Saunders, and J. Parkhill. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23. - PMC - PubMed
-
- Bourdoulous, S., and X. Nassif. 2006. Mechanisms of attachment and invasion, p. 257-272. In M. Frosch and M. C. Maiden (ed.), Handbook of meningococcal disease. Wiley-VCH, Weinheim, Germany.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

