AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes
- PMID: 20709902
- PMCID: PMC2950516
- DOI: 10.1128/JB.00711-10
AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes
Abstract
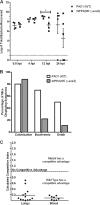
AmrZ is a putative ribbon-helix-helix (RHH) transcriptional regulator. RHH proteins utilize residues within the β-sheet for DNA binding, while the α-helices promote oligomerization. AmrZ is of interest due to its dual roles as a transcriptional activator and as a repressor, regulating genes encoding virulence factors associated with both chronic and acute Pseudomonas aeruginosa infection. In this study, cross-linking revealed that AmrZ forms oligomers in solution but that the amino terminus, containing an unordered region and a β-sheet, were not required for oligomerization. The first 12 unordered residues (extended amino terminus) contributed minimally to DNA binding. Mutagenesis of the AmrZ β-sheet demonstrated that residues 18, 20, and 22 were essential for DNA binding at both activation and repressor sites, suggesting that AmrZ utilizes a similar mechanism for binding to these sites. Mice infected with amrZ mutants exhibited reduced bacterial burden, morbidity, and mortality. Direct in vivo competition assays showed a 5-fold competitive advantage for the wild type over an isogenic amrZ mutant. Finally, the reduced infection phenotype of the amrZ-null strain was similar to that of a strain expressing a DNA-binding-deficient AmrZ variant, indicating that DNA binding and transcriptional regulation by AmrZ is responsible for the in vivo virulence defect. These recent infection data, along with previously identified AmrZ-regulated virulence factors, suggest the necessity of AmrZ transcriptional regulation for optimal virulence during acute infection.
Figures
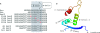





References
-
- Baynham, P. J., A. L. Brown, L. L. Hall, and D. J. Wozniak. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33:1069-1080. - PubMed
-
- Baynham, P. J., and D. J. Wozniak. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22:97-108. - PubMed
-
- Benanti, E. L., and P. T. Chivers. 2007. The N-terminal arm of the Helicobacter pylori Ni2+-dependent transcription factor NikR is required for specific DNA binding. J. Biol. Chem. 282:20365-20375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

