Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells
- PMID: 20711181
- PMCID: PMC3839080
- DOI: 10.1038/ncb2092
Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells
Abstract
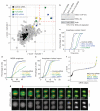
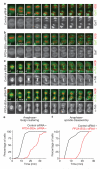
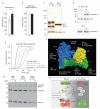
When vertebrate cells exit mitosis various cellular structures are re-organized to build functional interphase cells. This depends on Cdk1 (cyclin dependent kinase 1) inactivation and subsequent dephosphorylation of its substrates. Members of the protein phosphatase 1 and 2A (PP1 and PP2A) families can dephosphorylate Cdk1 substrates in biochemical extracts during mitotic exit, but how this relates to postmitotic reassembly of interphase structures in intact cells is not known. Here, we use a live-cell imaging assay and RNAi knockdown to screen a genome-wide library of protein phosphatases for mitotic exit functions in human cells. We identify a trimeric PP2A-B55alpha complex as a key factor in mitotic spindle breakdown and postmitotic reassembly of the nuclear envelope, Golgi apparatus and decondensed chromatin. Using a chemically induced mitotic exit assay, we find that PP2A-B55alpha functions downstream of Cdk1 inactivation. PP2A-B55alpha isolated from mitotic cells had reduced phosphatase activity towards the Cdk1 substrate, histone H1, and was hyper-phosphorylated on all subunits. Mitotic PP2A complexes co-purified with the nuclear transport factor importin-beta1, and RNAi depletion of importin-beta1 delayed mitotic exit synergistically with PP2A-B55alpha. This demonstrates that PP2A-B55alpha and importin-beta1 cooperate in the regulation of postmitotic assembly mechanisms in human cells.
Figures


References
-
- Morgan DO. The cell cycle: principles of control. Oxford University Press; 2007.
-
- Bollen M, Gerlich DW, Lesage B. Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 2009;19:531–541. - PubMed
-
- Trinkle-Mulcahy L, Lamond AI. Mitotic phosphatases: no longer silent partners. Curr. Opin. Cell Biol. 2006;18:623–631. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

