High-resolution mapping of protein sequence-function relationships
- PMID: 20711194
- PMCID: PMC2938879
- DOI: 10.1038/nmeth.1492
High-resolution mapping of protein sequence-function relationships
Abstract
We present a large-scale approach to investigate the functional consequences of sequence variation in a protein. The approach entails the display of hundreds of thousands of protein variants, moderate selection for activity and high-throughput DNA sequencing to quantify the performance of each variant. Using this strategy, we tracked the performance of >600,000 variants of a human WW domain after three and six rounds of selection by phage display for binding to its peptide ligand. Binding properties of these variants defined a high-resolution map of mutational preference across the WW domain; each position had unique features that could not be captured by a few representative mutations. Our approach could be applied to many in vitro or in vivo protein assays, providing a general means for understanding how protein function relates to sequence.
Figures
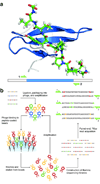



References
-
- Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr. Opin. Struct. Biol. 2007;17:481–487. - PubMed
-
- Matouschek A, Kellis JT, Jr, Serrano L, Fersht AR. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. - PubMed
-
- Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. - PubMed
-
- Levin AM, Weiss GA. Optimizing the affinity and specificity of proteins with molecular display. Mol. Biosyst. 2006;2:49–57. - PubMed
-
- Pal G, Kouadio JL, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for protein-protein interactions by rapid combinatorial scanning. J. Biol. Chem. 2006;281:22378–22385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

