Using DNA methylation patterns to infer tumor ancestry
- PMID: 20711251
- PMCID: PMC2918495
- DOI: 10.1371/journal.pone.0012002
Using DNA methylation patterns to infer tumor ancestry
Abstract
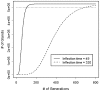
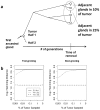
Background: Exactly how human tumors grow is uncertain because serial observations are impractical. One approach to reconstruct the histories of individual human cancers is to analyze the current genomic variation between its cells. The greater the variations, on average, the greater the time since the last clonal evolution cycle ("a molecular clock hypothesis"). Here we analyze passenger DNA methylation patterns from opposite sides of 12 primary human colorectal cancers (CRCs) to evaluate whether the variation (pairwise distances between epialleles) is consistent with a single clonal expansion after transformation.
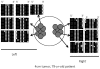
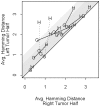
Methodology/principal findings: Data from 12 primary CRCs are compared to epigenomic data simulated under a single clonal expansion for a variety of possible growth scenarios. We find that for many different growth rates, a single clonal expansion can explain the population variation in 11 out of 12 CRCs. In eight CRCs, the cells from different glands are all equally distantly related, and cells sampled from the same tumor half appear no more closely related than cells sampled from opposite tumor halves. In these tumors, growth appears consistent with a single "symmetric" clonal expansion. In three CRCs, the variation in epigenetic distances was different between sides, but this asymmetry could be explained by a single clonal expansion with one region of a tumor having undergone more cell division than the other. The variation in one CRC was complex and inconsistent with a simple single clonal expansion.
Conclusions: Rather than a series of clonal expansion after transformation, these results suggest that the epigenetic variation of present-day cancer cells in primary CRCs can almost always be explained by a single clonal expansion.
Conflict of interest statement
Figures


Similar articles
-
High DNA methylation pattern intratumoral diversity implies weak selection in many human colorectal cancers.PLoS One. 2011;6(6):e21657. doi: 10.1371/journal.pone.0021657. Epub 2011 Jun 28. PLoS One. 2011. PMID: 21738754 Free PMC article.
-
Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers.Proc Natl Acad Sci U S A. 2009 Mar 24;106(12):4828-33. doi: 10.1073/pnas.0810276106. Epub 2009 Mar 4. Proc Natl Acad Sci U S A. 2009. PMID: 19261858 Free PMC article.
-
Many colorectal cancers are "flat" clonal expansions.Cell Cycle. 2009 Jul 15;8(14):2187-93. doi: 10.4161/cc.8.14.9151. Epub 2009 Jul 1. Cell Cycle. 2009. PMID: 19556889 Free PMC article. Review.
-
Modeling DNA methylation in a population of cancer cells.Stat Appl Genet Mol Biol. 2008;7(1):Article 18. doi: 10.2202/1544-6115.1374. Epub 2008 Jun 22. Stat Appl Genet Mol Biol. 2008. PMID: 18597664
-
Epigenetic alterations in colorectal cancer: the CpG island methylator phenotype.Histol Histopathol. 2013 May;28(5):585-95. doi: 10.14670/HH-28.585. Epub 2013 Jan 23. Histol Histopathol. 2013. PMID: 23341177 Review.
Cited by
-
Molecular tumor clocks to study the evolution of drug resistance.Mol Pharm. 2011 Dec 5;8(6):2050-4. doi: 10.1021/mp200256n. Epub 2011 Jul 12. Mol Pharm. 2011. PMID: 21732670 Free PMC article. Review.
-
Simulation framework for generating intratumor heterogeneity patterns in a cancer cell population.PLoS One. 2017 Sep 6;12(9):e0184229. doi: 10.1371/journal.pone.0184229. eCollection 2017. PLoS One. 2017. PMID: 28877206 Free PMC article.
-
The evolutionary theory of cancer: challenges and potential solutions.Nat Rev Cancer. 2024 Oct;24(10):718-733. doi: 10.1038/s41568-024-00734-2. Epub 2024 Sep 10. Nat Rev Cancer. 2024. PMID: 39256635 Free PMC article. Review.
-
High DNA methylation pattern intratumoral diversity implies weak selection in many human colorectal cancers.PLoS One. 2011;6(6):e21657. doi: 10.1371/journal.pone.0021657. Epub 2011 Jun 28. PLoS One. 2011. PMID: 21738754 Free PMC article.
-
Ancestral inference in tumors: how much can we know?J Theor Biol. 2014 Oct 21;359:136-45. doi: 10.1016/j.jtbi.2014.05.027. Epub 2014 Jun 5. J Theor Biol. 2014. PMID: 24907673 Free PMC article.
References
-
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. - PubMed
-
- Siegmund KD, Marjoram P, Shibata D. Modeling DNA methylation in a population of cancer cells. Stat Appl Genet Mol Biol. 2008;7:Article 18. - PubMed
-
- Anderson ARA, Chaplain MAJ, Rejniak KA. Single-Cell-Based Models in Biology and Medicine. Basel: Birkhauser-Verlag; 2007.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

