A genome-wide gene function prediction resource for Drosophila melanogaster
- PMID: 20711346
- PMCID: PMC2920829
- DOI: 10.1371/journal.pone.0012139
A genome-wide gene function prediction resource for Drosophila melanogaster
Abstract
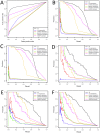
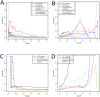
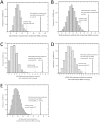
Predicting gene functions by integrating large-scale biological data remains a challenge for systems biology. Here we present a resource for Drosophila melanogaster gene function predictions. We trained function-specific classifiers to optimize the influence of different biological datasets for each functional category. Our model predicted GO terms and KEGG pathway memberships for Drosophila melanogaster genes with high accuracy, as affirmed by cross-validation, supporting literature evidence, and large-scale RNAi screens. The resulting resource of prioritized associations between Drosophila genes and their potential functions offers a guide for experimental investigations.
Conflict of interest statement
Figures


References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. - PubMed
-
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. - PubMed
-
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
-
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

