The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis
- PMID: 20711500
- PMCID: PMC2918508
- DOI: 10.1371/journal.pone.0012089
The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis
Abstract
Background: Bacillus anthracis, Francisella tularensis, and Yersinia pestis are bacterial pathogens that can cause anthrax, lethal acute pneumonic disease, and bubonic plague, respectively, and are listed as NIAID Category A priority pathogens for possible use as biological weapons. However, the interactions between human proteins and proteins in these bacteria remain poorly characterized leading to an incomplete understanding of their pathogenesis and mechanisms of immune evasion.
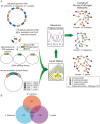
Methodology: In this study, we used a high-throughput yeast two-hybrid assay to identify physical interactions between human proteins and proteins from each of these three pathogens. From more than 250,000 screens performed, we identified 3,073 human-B. anthracis, 1,383 human-F. tularensis, and 4,059 human-Y. pestis protein-protein interactions including interactions involving 304 B. anthracis, 52 F. tularensis, and 330 Y. pestis proteins that are uncharacterized. Computational analysis revealed that pathogen proteins preferentially interact with human proteins that are hubs and bottlenecks in the human PPI network. In addition, we computed modules of human-pathogen PPIs that are conserved amongst the three networks. Functionally, such conserved modules reveal commonalities between how the different pathogens interact with crucial host pathways involved in inflammation and immunity.
Significance: These data constitute the first extensive protein interaction networks constructed for bacterial pathogens and their human hosts. This study provides novel insights into host-pathogen interactions.
Conflict of interest statement
Figures



Similar articles
-
Comparison of eleven commercially available rapid tests for detection of Bacillus anthracis, Francisella tularensis and Yersinia pestis.Lett Appl Microbiol. 2015 May;60(5):409-13. doi: 10.1111/lam.12392. Epub 2015 Feb 8. Lett Appl Microbiol. 2015. PMID: 25598285
-
Machine-Learning-Based Predictor of Human-Bacteria Protein-Protein Interactions by Incorporating Comprehensive Host-Network Properties.J Proteome Res. 2019 May 3;18(5):2195-2205. doi: 10.1021/acs.jproteome.9b00074. Epub 2019 Apr 22. J Proteome Res. 2019. PMID: 30983371
-
Bacillus anthracis, Francisella tularensis and Yersinia pestis. The most important bacterial warfare agents - review.Folia Microbiol (Praha). 2009;54(4):263-72. doi: 10.1007/s12223-009-0046-1. Epub 2009 Oct 14. Folia Microbiol (Praha). 2009. PMID: 19826916 Review.
-
Reevaluating limits of detection of 12 lateral flow immunoassays for the detection of Yersinia pestis, Francisella tularensis, and Bacillus anthracis spores using viable risk group-3 strains.J Appl Microbiol. 2021 Apr;130(4):1173-1180. doi: 10.1111/jam.14863. Epub 2020 Oct 2. J Appl Microbiol. 2021. PMID: 32970936
-
Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases.Nat Rev Microbiol. 2009 Nov;7(11):813-21. doi: 10.1038/nrmicro2219. Epub 2009 Oct 12. Nat Rev Microbiol. 2009. PMID: 19820723 Free PMC article. Review.
Cited by
-
Associations between HIV and human pathways revealed by protein-protein interactions and correlated gene expression profiles.PLoS One. 2012;7(3):e34240. doi: 10.1371/journal.pone.0034240. Epub 2012 Mar 27. PLoS One. 2012. PMID: 22479575 Free PMC article.
-
A Molecular Interaction Map of Klebsiella pneumoniae and Its Human Host Reveals Potential Mechanisms of Host Cell Subversion.Front Microbiol. 2021 Feb 18;12:613067. doi: 10.3389/fmicb.2021.613067. eCollection 2021. Front Microbiol. 2021. PMID: 33679637 Free PMC article.
-
Centrality in the host-pathogen interactome is associated with pathogen fitness during infection.Nat Commun. 2017 Jan 16;8:14092. doi: 10.1038/ncomms14092. Nat Commun. 2017. PMID: 28090086 Free PMC article.
-
Comparative interactomics for virus-human protein-protein interactions: DNA viruses versus RNA viruses.FEBS Open Bio. 2017 Jan 4;7(1):96-107. doi: 10.1002/2211-5463.12167. eCollection 2017 Jan. FEBS Open Bio. 2017. PMID: 28097092 Free PMC article.
-
A review on host-pathogen interactions: classification and prediction.Eur J Clin Microbiol Infect Dis. 2016 Oct;35(10):1581-99. doi: 10.1007/s10096-016-2716-7. Epub 2016 Jul 29. Eur J Clin Microbiol Infect Dis. 2016. PMID: 27470504 Review.
References
-
- Fukao T. Immune system paralysis by anthrax lethal toxin: the roles of innate and adaptive immunity. Lancet Infect Dis. 2004;4:166–170. - PubMed
-
- Fang H, Cordoba-Rodriguez R, Lankford CS, Frucht DM. Anthrax lethal toxin blocks MAPK kinase-dependent IL-2 production in CD4+ T cells. J Immunol. 2005;174:4966–4971. - PubMed
-
- Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, et al. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol. 2006;7:1066–1073. - PubMed
-
- Bosio CM, Bielefeldt-Ohmann H, Belisle JT. Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J Immunol. 2007;178:4538–4547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

