Searching for the Nik operon: how a ligand-responsive transcription factor hunts for its DNA binding site
- PMID: 20712334
- PMCID: PMC2934762
- DOI: 10.1021/bi100947k
Searching for the Nik operon: how a ligand-responsive transcription factor hunts for its DNA binding site
Abstract
Transcription factors regulate a wide variety of genes in the cell and play a crucial role in maintaining cellular homeostasis. A major unresolved issue is how transcription factors find their specific DNA binding sequence in the vast expanse of the cell and how they do so at rates that appear faster than the diffusion limit. Here, we relate an atomic-detail model that has been developed to describe the transcription factor NikR's mechanism of DNA binding to the broader theories of how transcription factors find their binding sites on DNA. NikR is the nickel regulatory transcription factor for many bacteria, and NikR from Escherichia coli is one of the best studied ligand-mediated transcription factors. For the E. coli NikR protein, there is a wide variety of structural, biochemical, and computational studies that provide significant insight into the NikR-DNA binding mechanism. We find that the two models, the atomic-level model for E. coli NikR and the cellular model for transcription factors in general, are in agreement, and the details laid out by the NikR system may lend additional credence to the current models for transcription factors searching for DNA.
Figures
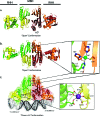
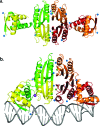


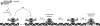

Similar articles
-
Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.Biochemistry. 2010 Sep 14;49(36):7830-8. doi: 10.1021/bi100923j. Biochemistry. 2010. PMID: 20704276 Free PMC article.
-
Structural basis of the metal specificity for nickel regulatory protein NikR.Biochemistry. 2008 Feb 19;47(7):1938-46. doi: 10.1021/bi702006h. Epub 2008 Jan 15. Biochemistry. 2008. PMID: 18193897 Free PMC article.
-
Potassium is critical for the Ni(II)-responsive DNA-binding activity of Escherichia coli NikR.J Am Chem Soc. 2010 Feb 10;132(5):1506-7. doi: 10.1021/ja909136h. J Am Chem Soc. 2010. PMID: 20088519
-
Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators.Biometals. 2005 Aug;18(4):413-28. doi: 10.1007/s10534-005-3716-8. Biometals. 2005. PMID: 16158234 Review.
-
MarR family transcription factors: dynamic variations on a common scaffold.Crit Rev Biochem Mol Biol. 2017 Dec;52(6):595-613. doi: 10.1080/10409238.2017.1344612. Epub 2017 Jul 3. Crit Rev Biochem Mol Biol. 2017. PMID: 28670937 Review.
Cited by
-
Allosteric control of metal-responsive transcriptional regulators in bacteria.J Biol Chem. 2020 Feb 7;295(6):1673-1684. doi: 10.1074/jbc.REV119.011444. Epub 2019 Dec 19. J Biol Chem. 2020. PMID: 31857375 Free PMC article. Review.
-
Metallochaperones and metalloregulation in bacteria.Essays Biochem. 2017 May 9;61(2):177-200. doi: 10.1042/EBC20160076. Print 2017 May 9. Essays Biochem. 2017. PMID: 28487396 Free PMC article. Review.
References
-
- Schreiter E. R.; Sintchak M. D.; Guo Y.; Chivers P. T.; Sauer R. T.; Drennan C. L. (2003) Crystal structure of nickel-responsive transcription factor NikR. Nat. Struct. Biol. 10, 794–799. - PubMed
-
- Chivers P. T.; Sauer R. T. (2002) NikR Repressor: High-Affinity Nickel Binding to the C-Terminal Domain Regulates Binding to Operator DNA. Chem. Biol. 9, 1141–1148. - PubMed
-
- Wang S. C.; Dias A. V.; Bloom S. L.; Zamble D. B. (2004) Selectivity of Metal Binding and Metal-Induced Stability of Escherichia coli NikR. Biochemistry 43, 10018–10028. - PubMed
-
- Bloom S. L.; Zamble D. B. (2004) Metal-Selective DNA-Binding Response of Escherichia coli NikR. Biochemistry 43, 10029–10038. - PubMed
-
- Navarro C.; Wu L. F.; Mandrand-Berthelot M.-A. (1993) The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9, 1181–1191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

