Characterization of hMTr1, a human Cap1 2'-O-ribose methyltransferase
- PMID: 20713356
- PMCID: PMC2963352
- DOI: 10.1074/jbc.M110.155283
Characterization of hMTr1, a human Cap1 2'-O-ribose methyltransferase
Abstract
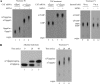
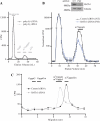
Cellular eukaryotic mRNAs are capped at their 5' ends with a 7-methylguanosine nucleotide, a structural feature that has been shown to be important for conferring mRNA stability, stimulating mRNA biogenesis (splicing, poly(A) addition, nucleocytoplasmic transport), and increasing translational efficiency. Whereas yeast mRNAs have no additional modifications to the cap, called cap0, higher eukaryotes are methylated at the 2'-O-ribose of the first or the first and second transcribed nucleotides, called cap1 and cap2, respectively. In the present study, we identify the methyltransferase responsible for cap1 formation in human cells, which we call hMTr1 (also known as FTSJD2 and ISG95). We show in vitro that hMTr1 catalyzes specific methylation of the 2'-O-ribose of the first nucleotide of a capped RNA transcript. Using siRNA-mediated knockdown of hMTr1 in HeLa cells, we demonstrate that hMTr1 is responsible for cap1 formation in vivo.
Figures



References
-
- Shatkin A. J. (1976) Cell 9, 645–653 - PubMed
-
- Gu M., Lima C. D. (2005) Curr. Opin. Struct. Biol. 15, 99–106 - PubMed
-
- Shuman S. (2002) Nat. Rev. Mol. Cell. Biol. 3, 619–625 - PubMed
-
- Ensinger M. J., Moss B. (1976) J. Biol. Chem. 251, 5283–5291 - PubMed
-
- Venkatesan S., Gershowitz A., Moss B. (1980) J. Biol. Chem. 255, 2829–2834 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

