Expanded roles of the Fanconi anemia pathway in preserving genomic stability
- PMID: 20713514
- PMCID: PMC2922498
- DOI: 10.1101/gad.1955310
Expanded roles of the Fanconi anemia pathway in preserving genomic stability
Abstract
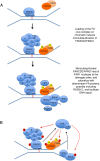
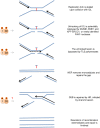
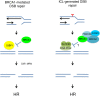
Studying rare human genetic diseases often leads to a better understanding of normal cellular functions. Fanconi anemia (FA), for example, has elucidated a novel DNA repair mechanism required for maintaining genomic stability and preventing cancer. The FA pathway, an essential tumor-suppressive pathway, is required for protecting the human genome from a specific type of DNA damage; namely, DNA interstrand cross-links (ICLs). In this review, we discuss the recent progress in the study of the FA pathway, such as the identification of new FANCM-binding partners and the identification of RAD51C and FAN1 (Fanconi-associated nuclease 1) as new FA pathway-related proteins. We also focus on the role of the FA pathway as a potential regulator of DNA repair choices in response to double-strand breaks, and its novel functions during the mitotic phase of the cell cycle.
Figures

References
-
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi Anemia. Mol Cell. 39: 25–35 - PubMed
-
- Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, van der Wal A, van der Valk M, Joenje H, te Riele H, et al. 2009. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet 18: 3484–3495 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
