Regulation of the autophagy protein LC3 by phosphorylation
- PMID: 20713600
- PMCID: PMC2928022
- DOI: 10.1083/jcb.201002108
Regulation of the autophagy protein LC3 by phosphorylation
Abstract
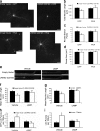
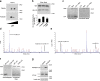
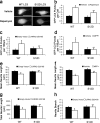
Macroautophagy is a major catabolic pathway that impacts cell survival, differentiation, tumorigenesis, and neurodegeneration. Although bulk degradation sustains carbon sources during starvation, autophagy contributes to shrinkage of differentiated neuronal processes. Identification of autophagy-related genes has spurred rapid advances in understanding the recruitment of microtubule-associated protein 1 light chain 3 (LC3) in autophagy induction, although braking mechanisms remain less understood. Using mass spectrometry, we identified a direct protein kinase A (PKA) phosphorylation site on LC3 that regulates its participation in autophagy. Both metabolic (rapamycin) and pathological (MPP(+)) inducers of autophagy caused dephosphorylation of endogenous LC3. The pseudophosphorylated LC3 mutant showed reduced recruitment to autophagosomes, whereas the nonphosphorylatable mutant exhibited enhanced puncta formation. Finally, autophagy-dependent neurite shortening induced by expression of a Parkinson disease-associated G2019S mutation in leucine-rich repeat kinase 2 was inhibited by dibutyryl-cyclic adenosine monophosphate, cytoplasmic expression of the PKA catalytic subunit, or the LC3 phosphorylation mimic. These data demonstrate a role for phosphorylation in regulating LC3 activity.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

