BAG-6 is essential for selective elimination of defective proteasomal substrates
- PMID: 20713601
- PMCID: PMC2928017
- DOI: 10.1083/jcb.200908092
BAG-6 is essential for selective elimination of defective proteasomal substrates
Abstract
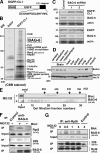
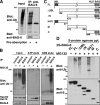
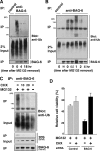
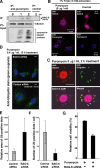
BAG-6/Scythe/BAT3 is a ubiquitin-like protein that was originally reported to be the product of a novel gene located within the human major histocompatibility complex, although the mechanisms of its function remain largely obscure. Here, we demonstrate the involvement of BAG-6 in the degradation of a CL1 model defective protein substrate in mammalian cells. We show that BAG-6 is essential for not only model substrate degradation but also the ubiquitin-mediated metabolism of newly synthesized defective polypeptides. Furthermore, our in vivo and in vitro analysis shows that BAG-6 interacts physically with puromycin-labeled nascent chain polypeptides and regulates their proteasome-mediated degradation. Finally, we show that knockdown of BAG-6 results in the suppressed presentation of MHC class I on the cell surface, a procedure known to be affected by the efficiency of metabolism of defective ribosomal products. Therefore, we propose that BAG-6 is necessary for ubiquitin-mediated degradation of newly synthesized defective polypeptides.
Figures


References
-
- Anton L.C., Yewdell J.W., Bennink J.R. 1997. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J. Immunol. 158:2535–2542 - PubMed
-
- Banerji J., Sands J., Strominger J.L., Spies T. 1990. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc. Natl. Acad. Sci. USA. 87:2374–2378 10.1073/pnas.87.6.2374 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

