Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster
- PMID: 20713603
- PMCID: PMC2928026
- DOI: 10.1083/jcb.201003001
Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster
Abstract
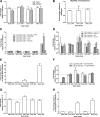
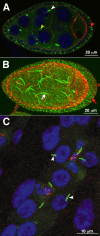
The discovery of large supramolecular complexes such as the purinosome suggests that subcellular organization is central to enzyme regulation. A screen of the yeast GFP strain collection to identify proteins that assemble into visible structures identified four novel filament systems comprised of glutamate synthase, guanosine diphosphate-mannose pyrophosphorylase, cytidine triphosphate (CTP) synthase, or subunits of the eIF2/2B translation factor complex. Recruitment of CTP synthase to filaments and foci can be modulated by mutations and regulatory ligands that alter enzyme activity, arguing that the assembly of these structures is related to control of CTP synthase activity. CTP synthase filaments are evolutionarily conserved and are restricted to axons in neurons. This spatial regulation suggests that these filaments have additional functions separate from the regulation of enzyme activity. The identification of four novel filaments greatly expands the number of known intracellular filament networks and has broad implications for our understanding of how cells organize biochemical activities in the cytoplasm.
Figures


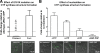
Similar articles
-
Characterization of filament-forming CTP synthases from Arabidopsis thaliana.Plant J. 2018 Oct;96(2):316-328. doi: 10.1111/tpj.14032. Epub 2018 Aug 31. Plant J. 2018. PMID: 30030857 Free PMC article.
-
Common regulatory control of CTP synthase enzyme activity and filament formation.Mol Biol Cell. 2014 Aug 1;25(15):2282-90. doi: 10.1091/mbc.E14-04-0912. Epub 2014 Jun 11. Mol Biol Cell. 2014. PMID: 24920825 Free PMC article.
-
Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.Elife. 2021 Nov 4;10:e73368. doi: 10.7554/eLife.73368. Elife. 2021. PMID: 34734801 Free PMC article.
-
Phospholipid synthesis in yeast: regulation by phosphorylation.Biochem Cell Biol. 2004 Feb;82(1):62-70. doi: 10.1139/o03-064. Biochem Cell Biol. 2004. PMID: 15052328 Review.
-
CTPS cytoophidia in Drosophila: distribution, regulation, and physiological roles.Exp Cell Res. 2025 Apr 15;447(2):114536. doi: 10.1016/j.yexcr.2025.114536. Epub 2025 Mar 22. Exp Cell Res. 2025. PMID: 40122502 Review.
Cited by
-
An expanded view of the eukaryotic cytoskeleton.Mol Biol Cell. 2013 Jun;24(11):1615-8. doi: 10.1091/mbc.E12-10-0732. Mol Biol Cell. 2013. PMID: 23722945 Free PMC article.
-
Differential Cytoophidium Assembly between Saccharomyces cerevisiae and Schizosaccharomyces pombe.Int J Mol Sci. 2024 Sep 19;25(18):10092. doi: 10.3390/ijms251810092. Int J Mol Sci. 2024. PMID: 39337578 Free PMC article.
-
Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth.Genome Res. 2012 Dec;22(12):2478-88. doi: 10.1101/gr.142802.112. Epub 2012 Jul 18. Genome Res. 2012. PMID: 22813931 Free PMC article.
-
The Need for Speed: Run-On Oligomer Filament Formation Provides Maximum Speed with Maximum Sequestration of Activity.J Virol. 2019 Feb 19;93(5):e01647-18. doi: 10.1128/JVI.01647-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30518649 Free PMC article.
-
Characterization of filament-forming CTP synthases from Arabidopsis thaliana.Plant J. 2018 Oct;96(2):316-328. doi: 10.1111/tpj.14032. Epub 2018 Aug 31. Plant J. 2018. PMID: 30030857 Free PMC article.
References
-
- Aronow B., Ullman B. 1987. In situ regulation of mammalian CTP synthetase by allosteric inhibition. J. Biol. Chem. 262:5106–5112 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

