The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia
- PMID: 20713618
- PMCID: PMC2949043
- DOI: 10.1104/pp.110.163949
The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia
Abstract
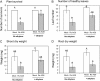
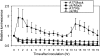
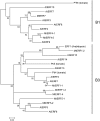
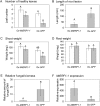
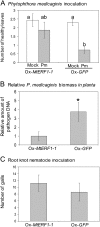
The fungal necrotrophic pathogen Rhizoctonia solani is a significant constraint to a range of crops as diverse as cereals, canola, and legumes. Despite wide-ranging germplasm screens in many of these crops, no strong genetic resistance has been identified, suggesting that alternative strategies to improve resistance are required. In this study, we characterize moderate resistance to R. solani anastomosis group 8 identified in Medicago truncatula. The activity of the ethylene- and jasmonate-responsive GCC box promoter element was associated with moderate resistance, as was the induction of the B-3 subgroup of ethylene response transcription factors (ERFs). Genes of the B-1 subgroup showed no significant response to R. solani infection. Overexpression of a B-3 ERF, MtERF1-1, in Medicago roots increased resistance to R. solani as well as an oomycete root pathogen, Phytophthora medicaginis, but not root knot nematode. These results indicate that targeting specific regulators of ethylene defense may enhance resistance to an important subset of root pathogens. We also demonstrate that overexpression of MtERF1-1 enhances disease resistance without apparent impact on nodulation in the A17 background, while overexpression in sickle reduced the hypernodulation phenotype. This suggests that under normal regulation of nodulation, enhanced resistance to root diseases can be uncoupled from symbiotic plant-microbe interactions in the same tissue and that ethylene/ERF regulation of nodule number is distinct from the defenses regulated by B-3 ERFs. Furthermore, unlike the stunted phenotype previously described for Arabidopsis (Arabidopsis thaliana) ubiquitously overexpressing B-3 ERFs, overexpression of MtERF1-1 in M. truncatula roots did not show adverse effects on plant development.
Figures



References
-
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. (2004a) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 - PMC - PubMed
-
- Anderson NA. (1982) The genetics and pathology of Rhizoctonia solani. Annu Rev Phytopathol 20: 329–347
-
- Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. (2004b) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol 19: 535–544 - PubMed
-
- Barbetti MJ, Riley IT, You MP, Li H, Sivasithamparam K. (2006) The association of necrotrophic fungal pathogens and plant parasitic nematodes with the loss of productivity of annual medic-based pastures in Australia and options for their management. Australas Plant Pathol 35: 691–706
-
- Berrocal-Lobo M, Molina A. (2004) ETHYLENE RESPONSE FACTOR 1 mediates Arabidopsis resistance to the soil-borne fungus Fusarium oxysporum. Mol Plant Microbe Interact 17: 673–770 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

