Tracking, tuning, and terminating microbial physiology using synthetic riboregulators
- PMID: 20713708
- PMCID: PMC2936621
- DOI: 10.1073/pnas.1009747107
Tracking, tuning, and terminating microbial physiology using synthetic riboregulators
Abstract
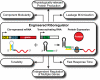
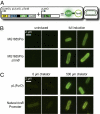
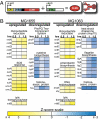
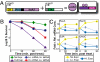
The development of biomolecular devices that interface with biological systems to reveal new insights and produce novel functions is one of the defining goals of synthetic biology. Our lab previously described a synthetic, riboregulator system that affords for modular, tunable, and tight control of gene expression in vivo. Here we highlight several experimental advantages unique to this RNA-based system, including physiologically relevant protein production, component modularity, leakage minimization, rapid response time, tunable gene expression, and independent regulation of multiple genes. We demonstrate this utility in four sets of in vivo experiments with various microbial systems. Specifically, we show that the synthetic riboregulator is well suited for GFP fusion protein tracking in wild-type cells, tight regulation of toxic protein expression, and sensitive perturbation of stress response networks. We also show that the system can be used for logic-based computing of multiple, orthogonal inputs, resulting in the development of a programmable kill switch for bacteria. This work establishes a broad, easy-to-use synthetic biology platform for microbiology experiments and biotechnology applications.
Conflict of interest statement
Conflict of interest statement: D.J.D., F.J.I, C.R.C., and J.J.C. have a patent for the riboregulator system pending.
Figures
References
-
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. - PubMed
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. - PubMed
-
- Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

