PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair
- PMID: 20713735
- PMCID: PMC2941292
- DOI: 10.1073/pnas.1010662107
PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair
Abstract
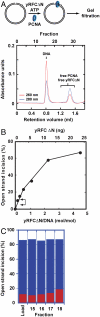
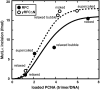
MutLα (MLH1-PMS2) is a latent endonuclease that is activated in a mismatch-, MutSα-, proliferating cell nuclear antigen (PCNA)-, replication factor C (RFC)-, and ATP-dependent manner, with nuclease action directed to the heteroduplex strand that contains a preexisting break. RFC depletion experiments and use of linear DNAs indicate that RFC function in endonuclease activation is limited to PCNA loading. Whereas nicked circular heteroduplex DNA is a good substrate for PCNA loading and for endonuclease activation on the incised strand, covalently closed, relaxed circular DNA is a poor substrate for both reactions. However, covalently closed supercoiled or bubble-containing relaxed heteroduplexes, which do support PCNA loading, also support MutLα activation, but in this case cleavage strand bias is largely abolished. Based on these findings we suggest that PCNA has two roles in MutLα function: The clamp is required for endonuclease activation, an effect that apparently involves interaction of the two proteins, and by virtue of its loading orientation, PCNA determines the strand direction of MutLα incision. These results also provide a potential mechanism for activation of mismatch repair on nonreplicating DNA, an effect that may have implications for the somatic phase of triplet repeat expansion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


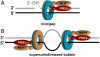
Comment in
-
PCNA and MutLα: partners in crime in triplet repeat expansion?Proc Natl Acad Sci U S A. 2010 Sep 21;107(38):16409-10. doi: 10.1073/pnas.1011692107. Epub 2010 Sep 7. Proc Natl Acad Sci U S A. 2010. PMID: 20823236 Free PMC article. No abstract available.
References
-
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. - PubMed
-
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. - PubMed
-
- Dzantiev L, et al. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

