Cutting edge: proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes
- PMID: 20713892
- PMCID: PMC3104018
- DOI: 10.4049/jimmunol.1001512
Cutting edge: proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes
Abstract
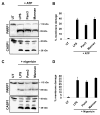
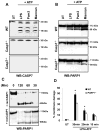
Caspase-mediated cleavage of the DNA damage sensor poly(ADP-ribose) polymerase 1 (PARP1) is a hallmark of apoptosis. However, it remains unclear whether PARP1 is processed during pyroptosis, a specialized cell-death program that occurs upon activation of caspase-1 in inflammasome complexes. In this article, we show that activation of the Nlrp3 and Nlrc4 inflammasomes induces processing of full-length PARP1 into a fragment of 89 kDa in a stimulus-dependent manner. Macrophages deficient for caspase-1 and those lacking the inflammasome adaptors Nlrp3, Nlrc4, and ASC were highly resistant to cleavage, whereas macrophages lacking the downstream inflammasome effector caspase-7 were partially protected. A modest, but statistically significant, reduction in Nlrp3 inflammasome-induced pyroptosis was observed in PARP1 knockout macrophages. Thus, protease-mediated inactivation of PARP1 is a shared feature of apoptotic, necrotic, and pyroptotic cells.
Figures

Similar articles
-
Differential regulation of caspase-1 activation via NLRP3/NLRC4 inflammasomes mediated by aerolysin and type III secretion system during Aeromonas veronii infection.J Immunol. 2010 Dec 1;185(11):7077-84. doi: 10.4049/jimmunol.1002165. Epub 2010 Oct 29. J Immunol. 2010. PMID: 21037094
-
Inflammasome-activated caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-κB target genes.Mol Cell. 2012 Apr 27;46(2):200-11. doi: 10.1016/j.molcel.2012.02.016. Epub 2012 Mar 29. Mol Cell. 2012. PMID: 22464733
-
Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes.J Clin Immunol. 2010 Sep;30(5):693-702. doi: 10.1007/s10875-010-9425-2. Epub 2010 May 20. J Clin Immunol. 2010. PMID: 20490635 Free PMC article.
-
Poly(ADP-ribose) Polymerase (PARP) and PARP Inhibitors: Mechanisms of Action and Role in Cardiovascular Disorders.Cardiovasc Toxicol. 2018 Dec;18(6):493-506. doi: 10.1007/s12012-018-9462-2. Cardiovasc Toxicol. 2018. PMID: 29968072 Review.
-
Regulation and Function of the Nucleotide Binding Domain Leucine-Rich Repeat-Containing Receptor, Pyrin Domain-Containing-3 Inflammasome in Lung Disease.Am J Respir Cell Mol Biol. 2016 Feb;54(2):151-60. doi: 10.1165/rcmb.2015-0231TR. Am J Respir Cell Mol Biol. 2016. PMID: 26418144 Free PMC article. Review.
Cited by
-
Great balls of fire: activation and signalling of inflammatory caspases.Biochem Soc Trans. 2021 Jun 30;49(3):1311-1324. doi: 10.1042/BST20200986. Biochem Soc Trans. 2021. PMID: 34060593 Free PMC article. Review.
-
The PANoptosome: A Deadly Protein Complex Driving Pyroptosis, Apoptosis, and Necroptosis (PANoptosis).Front Cell Infect Microbiol. 2020 Jun 3;10:238. doi: 10.3389/fcimb.2020.00238. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32582562 Free PMC article. Review.
-
Innate immune inflammatory cell death: PANoptosis and PANoptosomes in host defense and disease.Eur J Immunol. 2023 Nov;53(11):e2250235. doi: 10.1002/eji.202250235. Epub 2023 Mar 3. Eur J Immunol. 2023. PMID: 36782083 Free PMC article. Review.
-
Nicotinamide Augments the Anti-Inflammatory Properties of Resveratrol through PARP1 Activation.Sci Rep. 2019 Jul 15;9(1):10219. doi: 10.1038/s41598-019-46678-8. Sci Rep. 2019. PMID: 31308445 Free PMC article.
-
Poly-ADP-ribosylation-mediated degradation of ARTD1 by the NLRP3 inflammasome is a prerequisite for osteoclast maturation.Cell Death Dis. 2016 Mar 24;7(3):e2153. doi: 10.1038/cddis.2016.58. Cell Death Dis. 2016. PMID: 27010854 Free PMC article.
References
-
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004 Aug;26( 8):882–893. - PubMed
-
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005 Sep 1;19( 17):1951–1967. - PubMed
-
- Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004 May 6;23( 21):3872–3882. - PubMed
-
- Tewari M, Quan LT, O’Rourke K, Desnoyers S, Zeng Z, Beidler DR, et al. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995 Jun 2;81( 5):801–809. - PubMed
-
- Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995 Jul 6;376( 6535):37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

