Effects of time point measurement on the reconstruction of gene regulatory networks
- PMID: 20714301
- PMCID: PMC6257779
- DOI: 10.3390/molecules15085354
Effects of time point measurement on the reconstruction of gene regulatory networks
Abstract
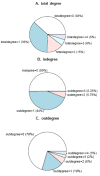
With the availability of high-throughput gene expression data in the post-genomic era, reconstruction of gene regulatory networks has become a hot topic. Regulatory networks have been intensively studied over the last decade and many software tools are currently available. However, the impact of time point selection on network reconstruction is often underestimated. In this paper we apply the Dynamic Bayesian network (DBN) to construct the Arabidopsis gene regulatory networks by analyzing the time-series gene microarray data. In order to evaluate the impact of time point measurement on network reconstruction, we deleted time points one by one to yield 11 distinct groups of incomplete time series. Then the gene regulatory networks constructed based on complete and incomplete data series are compared in terms of statistics at different levels. Two time points are found to play a significant role in the Arabidopsis gene regulatory networks. Pathway analysis of significant nodes revealed three key regulatory genes. In addition, important regulations between genes, which were insensitive to the time point measurement, were also identified.
Figures




References
-
- Kato M., Tsunoda T., Takagi T. Inferring genetic networks from DNA microarray data by multiple regression analysis. Genome Inform. Ser. Workshop Genome Inform. 2000;11:118–128. - PubMed
-
- Chen T., He H.L., Church G.M. Modeling gene expression with differential equations. Pac. Symp. Biocomput. 1999:29–40. - PubMed
-
- de Hoon M.J., Imoto S., Kobayashi K., Ogasawara N., Miyano S. Inferring gene regulatory networks from time-ordered gene expression data of Bacillus subtilis using differential equations. Pac. Symp. Biocomput. 2003:17–28. - PubMed
-
- Basso K., Margolin A.A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. - PubMed
-
- Liang S., Fuhrman S., Somogyi R. Reveal, a general reverse engineering algorithm for inference of genetic network architectures. Pac. Symp. Biocomput. 1998:18–29. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

