Molecular evolution of functional nucleic acids with chemical modifications
- PMID: 20714306
- PMCID: PMC6257756
- DOI: 10.3390/molecules15085423
Molecular evolution of functional nucleic acids with chemical modifications
Abstract
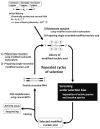
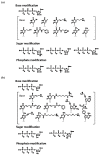
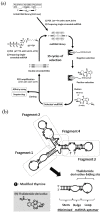
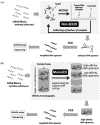
Nucleic acids are attractive materials for creating functional molecules that have applications as catalysts, specific binders, and molecular switches. Nucleic acids having such functions can be obtained by random screening, typically using in vitro selection methods. These methods have helped explore the potential abilities of nucleic acids and steadily contributed to their evolution, i.e., creation of RNA/DNA enzymes, aptamers, and aptazymes. Chemical modification would be a key means to further increase their performance, e.g., expansion of function diversity, enhancement of activity, and improvement of biostability for biological use. Indeed, in the past two decades, random screening involving chemical modification, post-SELEX chemical modification, and rational design methods have been advanced, and combining and integrating these methods may produce a new class of functional nucleic acids. This review focuses on the effectiveness of chemical modifications on the evolution of nucleic acids as functional molecules and the outlook for related technologies.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

