Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits
- PMID: 20714348
- PMCID: PMC2920848
- DOI: 10.1371/journal.pgen.1001058
Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits
Abstract
Mitochondrial dysfunction has been observed in skeletal muscle of people with diabetes and insulin-resistant individuals. Furthermore, inherited mutations in mitochondrial DNA can cause a rare form of diabetes. However, it is unclear whether mitochondrial dysfunction is a primary cause of the common form of diabetes. To date, common genetic variants robustly associated with type 2 diabetes (T2D) are not known to affect mitochondrial function. One possibility is that multiple mitochondrial genes contain modest genetic effects that collectively influence T2D risk. To test this hypothesis we developed a method named Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTA; http://www.broadinstitute.org/mpg/magenta). MAGENTA, in analogy to Gene Set Enrichment Analysis, tests whether sets of functionally related genes are enriched for associations with a polygenic disease or trait. MAGENTA was specifically designed to exploit the statistical power of large genome-wide association (GWA) study meta-analyses whose individual genotypes are not available. This is achieved by combining variant association p-values into gene scores and then correcting for confounders, such as gene size, variant number, and linkage disequilibrium properties. Using simulations, we determined the range of parameters for which MAGENTA can detect associations likely missed by single-marker analysis. We verified MAGENTA's performance on empirical data by identifying known relevant pathways in lipid and lipoprotein GWA meta-analyses. We then tested our mitochondrial hypothesis by applying MAGENTA to three gene sets: nuclear regulators of mitochondrial genes, oxidative phosphorylation genes, and approximately 1,000 nuclear-encoded mitochondrial genes. The analysis was performed using the most recent T2D GWA meta-analysis of 47,117 people and meta-analyses of seven diabetes-related glycemic traits (up to 46,186 non-diabetic individuals). This well-powered analysis found no significant enrichment of associations to T2D or any of the glycemic traits in any of the gene sets tested. These results suggest that common variants affecting nuclear-encoded mitochondrial genes have at most a small genetic contribution to T2D susceptibility.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 for SNP i. Gene boundaries (vertical dashed lines) are defined here as predetermined physical distances added upstream and downstream to the most extreme transcript start and end sites of the gene (red arrow), respectively. Linkage-based distances can also be used. Each gene is assigned a set of SNPs that fall in its gene region boundaries. Two genes are shown for simplicity. (B) Step 2: Score genes based on their local SNP
for SNP i. Gene boundaries (vertical dashed lines) are defined here as predetermined physical distances added upstream and downstream to the most extreme transcript start and end sites of the gene (red arrow), respectively. Linkage-based distances can also be used. Each gene is assigned a set of SNPs that fall in its gene region boundaries. Two genes are shown for simplicity. (B) Step 2: Score genes based on their local SNP  . Here the most significant
. Here the most significant  of all SNPs i that lie within the extended gene boundaries is assigned to each gene g in the genome (
of all SNPs i that lie within the extended gene boundaries is assigned to each gene g in the genome ( ). (C) Step 3: Correct for confounding effects on the gene score,
). (C) Step 3: Correct for confounding effects on the gene score,  in the absence of genotype data. In this study we used step-wise multivariate linear regression analysis to regress out of
in the absence of genotype data. In this study we used step-wise multivariate linear regression analysis to regress out of  the confounding effects of several physical and genetic properties of genes (listed in Table 1);
the confounding effects of several physical and genetic properties of genes (listed in Table 1);  refers to the corrected gene p-value for gene g. In cases where two genes are assigned the same best SNP p-value,
refers to the corrected gene p-value for gene g. In cases where two genes are assigned the same best SNP p-value,  tends to be more significant for small genes than for large genes. (D) Step 4: Calculate a gene set enrichment p-value for each biological pathway or gene set of interest. We used a non-parametric statistical test to test whether
tends to be more significant for small genes than for large genes. (D) Step 4: Calculate a gene set enrichment p-value for each biological pathway or gene set of interest. We used a non-parametric statistical test to test whether  for all genes in gene set gs are enriched for highly ranked gene scores more than would be expected by chance, compared to randomly sampled gene sets of identical size from the genome.
for all genes in gene set gs are enriched for highly ranked gene scores more than would be expected by chance, compared to randomly sampled gene sets of identical size from the genome.  refers to the nominal gene set enrichment p-value for gene set gs.
refers to the nominal gene set enrichment p-value for gene set gs.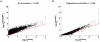
 was evaluated against permutation analysis correction, since the latter corrects for all confounders without requiring a priori knowledge of them. T2D gene association p-values were plotted for all genes g in the genome (A) before gene score adjustment (
was evaluated against permutation analysis correction, since the latter corrects for all confounders without requiring a priori knowledge of them. T2D gene association p-values were plotted for all genes g in the genome (A) before gene score adjustment ( ) and (B) after correction for confounders using regression analysis (
) and (B) after correction for confounders using regression analysis ( ), as a function of corrected gene p-values using phenotype permutation analysis (
), as a function of corrected gene p-values using phenotype permutation analysis ( ). The Diabetes Genetics Initiative (DGI) GWA study was used for the analysis, since we had access to all individuals' genotypes.
). The Diabetes Genetics Initiative (DGI) GWA study was used for the analysis, since we had access to all individuals' genotypes.  is the association p-value of the best regional SNP for gene g before correction (y-axis in A). To compute
is the association p-value of the best regional SNP for gene g before correction (y-axis in A). To compute  (y-axis in B), step-wise multivariate linear regression analysis was applied to
(y-axis in B), step-wise multivariate linear regression analysis was applied to  against the first four confounders listed in Table 1 (this approach does not require genotype data). The Pearson's correlation coefficient (calculated between p-value vectors before log transformation) increased significantly following the regression-based correction (from r = 0.69 to r = 0.95). The spread around the diagonal (red line) also decreased following the regression correction (from a coefficient of variation (mean/std) of 1.13 to 0.56). The minimum
against the first four confounders listed in Table 1 (this approach does not require genotype data). The Pearson's correlation coefficient (calculated between p-value vectors before log transformation) increased significantly following the regression-based correction (from r = 0.69 to r = 0.95). The spread around the diagonal (red line) also decreased following the regression correction (from a coefficient of variation (mean/std) of 1.13 to 0.56). The minimum  is 10−4 as the p-values were calculated based on 1,000 permutations for genes with
is 10−4 as the p-values were calculated based on 1,000 permutations for genes with  , and 10,000 permutations for genes with
, and 10,000 permutations for genes with  . Some of the variation in the low p-value tail is due to having done only 10,000 permutations (
. Some of the variation in the low p-value tail is due to having done only 10,000 permutations ( ), and some to limitations of the linear regression method. Note that the four dots in (A) with
), and some to limitations of the linear regression method. Note that the four dots in (A) with  contain ten overlapping dots that refer to four sets of 2–3 genes, each set assigned the same
contain ten overlapping dots that refer to four sets of 2–3 genes, each set assigned the same  . Gene association p-values are plotted on a −log10(p-value) scale.
. Gene association p-values are plotted on a −log10(p-value) scale.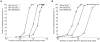
 vectors from case/control permutations of the DGI study were used as the background association values. Simulations were repeated 1,000 times for each unique set of parameters. Power was calculated as the fraction of times the simulated gene set received a
vectors from case/control permutations of the DGI study were used as the background association values. Simulations were repeated 1,000 times for each unique set of parameters. Power was calculated as the fraction of times the simulated gene set received a  <0.01. For specificity estimations we used SNPs with no effect size, sampled from a null distribution that assumes no association. The false positive rate of the method (1-specificity) was comparable to the p-value cutoff used (0.3–1.7%). Note the x-axis in both panels is on a log10 scale.
<0.01. For specificity estimations we used SNPs with no effect size, sampled from a null distribution that assumes no association. The false positive rate of the method (1-specificity) was comparable to the p-value cutoff used (0.3–1.7%). Note the x-axis in both panels is on a log10 scale.References
-
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. - DOI - PubMed
-
- Dumas J, Simard G, Flamment M, Ducluzeau P, Ritz P. Is skeletal muscle mitochondrial dysfunction a cause or an indirect consequence of insulin resistance in humans? Diabetes Metab. 2009;35:159–167. doi: 10.1016/j.diabet.2009.02.002. - DOI - PubMed
-
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. - DOI - PMC - PubMed
-
- Jin W, Patti M. Genetic determinants and molecular pathways in the pathogenesis of Type 2 diabetes. Clin Sci. 2009;116:99–111. doi: 10.1042/CS20080090. - DOI - PubMed
-
- Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

