Cloning and comparative studies of seaweed trehalose-6-phosphate synthase genes
- PMID: 20714424
- PMCID: PMC2920543
- DOI: 10.3390/md8072065
Cloning and comparative studies of seaweed trehalose-6-phosphate synthase genes
Abstract
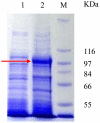
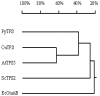
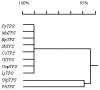
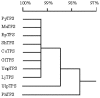
The full-length cDNA sequence (3219 base pairs) of the trehalose-6-phosphate synthase gene of Porphyra yezoensis (PyTPS) was isolated by RACE-PCR and deposited in GenBank (NCBI) with the accession number AY729671. PyTPS encodes a protein of 908 amino acids before a stop codon, and has a calculated molecular mass of 101,591 Daltons. The PyTPS protein consists of a TPS domain in the N-terminus and a putative TPP domain at the C-terminus. Homology alignment for PyTPS and the TPS proteins from bacteria, yeast and higher plants indicated that the most closely related sequences to PyTPS were those from higher plants (OsTPS and AtTPS5), whereas the most distant sequence to PyTPS was from bacteria (EcOtsAB). Based on the identified sequence of the PyTPS gene, PCR primers were designed and used to amplify the TPS genes from nine other seaweed species. Sequences of the nine obtained TPS genes were deposited in GenBank (NCBI). All 10 TPS genes encoded peptides of 908 amino acids and the sequences were highly conserved both in nucleotide composition (>94%) and in amino acid composition (>96%). Unlike the TPS genes from some other plants, there was no intron in any of the 10 isolated seaweed TPS genes.
Keywords: RACE-PCR; comparative genomics of TPS genes; gene cloning; seaweed; trehalose-6-phosphate synthase gene.
Figures



References
-
- Zhang XC. Introduction. In: Zhang XC, Qin S, Ma JH, Xu P, editors. The Genetics of Marine Algae. Chinese Agriculture Press; Beijing, China: 2005. pp. 1–14. (In Chinese with English title and content list)
-
- Chan CX, Ho CL, Phang SM. Trends in seaweed research. Trends Plant Sci. 2006;11:165–166. - PubMed
-
- Goeme A. A small introduction on bioenergy and alage: the new biofuel? [(accessed on 14 June 2008)]. Available online: http://algaetobioenergy.wordpress.com.
-
- Reith H, Huijgen W, Hal JV, Lenstra J. Seaweed potential in the Netherlands. [(accessed on 1 October 2009)]. ECN report, 2 June 2009. Available online: http://www.supergen-bioenergy.net/Resources/user/docs/new/Hans%20Reith-S....
-
- Qin S, Jiang P, Tseng CK. Molecular biotechnology of marine algae in China. Hydrobiologia. 2004;512:21–26.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
