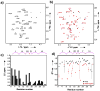
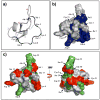
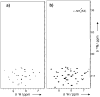
Backbone dynamics of cyclotide MCoTI-I free and complexed with trypsin
- PMID: 20715250
- PMCID: PMC2944905
- DOI: 10.1002/anie.201002906
Backbone dynamics of cyclotide MCoTI-I free and complexed with trypsin
Erratum in
- Angew Chem Int Ed Engl. 2011 Jul 25;50(31):6948-9
Figures
Similar articles
-
Computational analysis of the MCoTI-II plant defence knottin reveals a novel intermediate conformation that facilitates trypsin binding.Sci Rep. 2016 Mar 15;6:23174. doi: 10.1038/srep23174. Sci Rep. 2016. PMID: 26975976 Free PMC article.
-
Cyclotides, a versatile ultrastable micro-protein scaffold for biotechnological applications.Bioorg Med Chem Lett. 2017 Dec 1;27(23):5089-5099. doi: 10.1016/j.bmcl.2017.10.051. Epub 2017 Oct 21. Bioorg Med Chem Lett. 2017. PMID: 29110985 Free PMC article. Review.
-
Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor.J Biol Chem. 2013 Dec 13;288(50):36141-8. doi: 10.1074/jbc.M113.528240. Epub 2013 Oct 29. J Biol Chem. 2013. PMID: 24169696 Free PMC article.
-
Knots in rings. The circular knotted protein Momordica cochinchinensis trypsin inhibitor-II folds via a stable two-disulfide intermediate.J Biol Chem. 2006 Mar 24;281(12):8224-32. doi: 10.1074/jbc.M513399200. Epub 2006 Jan 23. J Biol Chem. 2006. PMID: 16547012
-
Oxidative folding of the cystine knot motif in cyclotide proteins.Protein Pept Lett. 2005 Feb;12(2):147-52. doi: 10.2174/0929866053005863. Protein Pept Lett. 2005. PMID: 15723640 Review.
Cited by
-
Chemical synthesis, backbone cyclization and oxidative folding of cystine-knot peptides: promising scaffolds for applications in drug design.Molecules. 2012 Oct 24;17(11):12533-52. doi: 10.3390/molecules171112533. Molecules. 2012. PMID: 23095896 Free PMC article. Review.
-
Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide-protein interactions.Angew Chem Int Ed Engl. 2013 Mar 11;52(11):3126-31. doi: 10.1002/anie.201209219. Epub 2013 Jan 15. Angew Chem Int Ed Engl. 2013. PMID: 23322720 Free PMC article. No abstract available.
-
Intein applications: from protein purification and labeling to metabolic control methods.J Biol Chem. 2014 May 23;289(21):14512-9. doi: 10.1074/jbc.R114.552653. Epub 2014 Apr 2. J Biol Chem. 2014. PMID: 24700459 Free PMC article. Review.
-
Design of a MCoTI-Based Cyclotide with Angiotensin (1-7)-Like Activity.Molecules. 2016 Jan 26;21(2):152. doi: 10.3390/molecules21020152. Molecules. 2016. PMID: 26821010 Free PMC article.
-
The Potential of the Cyclotide Scaffold for Drug Development.Biomedicines. 2019 Apr 19;7(2):31. doi: 10.3390/biomedicines7020031. Biomedicines. 2019. PMID: 31010257 Free PMC article. Review.
References
-
- Daly NL, Rosengren KJ, Craik DJ. Adv Drug Deliv Rev. 2009;61:918. - PubMed
-
- Craik DJ, Simonsen S, Daly NL. Curr Opin Drug Discov Devel. 2002;5:251. - PubMed
-
- Craik DJ, Cemazar M, Wang CK, Daly NL. Biopolymers. 2006;84:250. - PubMed
-
- Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D. Biochemistry. 2000;39:5722. - PubMed
-
- Heitz A, Hernandez JF, Gagnon J, Hong TT, Pham TT, Nguyen TM, Le-Nguyen D, Chiche L. Biochemistry. 2001;40:7973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

