The diheme cytochrome c(4) from Vibrio cholerae is a natural electron donor to the respiratory cbb(3) oxygen reductase
- PMID: 20715760
- PMCID: PMC2932843
- DOI: 10.1021/bi1004574
The diheme cytochrome c(4) from Vibrio cholerae is a natural electron donor to the respiratory cbb(3) oxygen reductase
Abstract
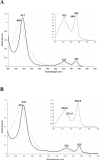
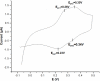
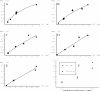
The respiratory chain of Vibrio cholerae contains three bd-type quinol oxygen reductases as well as one cbb(3) oxygen reductase. The cbb(3) oxygen reductase has been previously isolated and characterized; however, the natural mobile electron donor(s) that shuttles electrons between the bc(1) complex and the cbb(3) oxygen reductase is not known. The most likely candidates are the diheme cytochrome c(4) and monoheme cytochrome c(5), which have been previously shown to be present in the periplasm of aerobically grown cultures of V. cholerae. Both cytochromes c(4) and c(5) from V. cholerae have been cloned and expressed heterologously in Escherichia coli. It is shown that reduced cytochrome c(4) is a substrate for the purified cbb(3) oxygen reductase and can support steady state oxygen reductase activity of at least 300 e(-1)/s. In contrast, reduced cytochrome c(5) is not a good substrate for the cbb(3) oxygen reductase. Surprisingly, the dependence of the oxygen reductase activity on the concentration of cytochrome c(4) does not exhibit saturation. Global spectroscopic analysis of the time course of the oxidation of cytochrome c(4) indicates that the apparent lack of saturation is due to the strong dependence of K(M) and V(max) on the concentration of oxidized cytochrome c(4). Whether this is an artifact of the in vitro assay or has physiological significance remains unknown. Cyclic voltammetry was used to determine that the midpoint potentials of the two hemes in cytochrome c(4) are 240 and 340 mV (vs standard hydrogen electrode), similar to the electrochemical properties of other c(4)-type cytochromes. Genomic analysis shows a strong correlation between the presence of a c(4)-type cytochrome and a cbb(3) oxygen reductase within the beta- and gamma-proteobacterial clades, suggesting that cytochrome c(4) is the likely natural electron donor to the cbb(3) oxygen reductases within these organisms. These would include the beta-proteobacteria Neisseria meningitidis and Neisseria gonnorhoeae, in which the cbb(3) oxygen reductases are the only terminal oxidases in their respiratory chains, and the gamma-proteobacterium Pseudomonas stutzeri.
Figures


Similar articles
-
Functional importance of a pair of conserved glutamic acid residues and of Ca(2+) binding in the cbb(3)-type oxygen reductases from Rhodobacter sphaeroides and Vibrio cholerae.Biochemistry. 2012 Sep 18;51(37):7290-6. doi: 10.1021/bi3006847. Epub 2012 Sep 4. Biochemistry. 2012. PMID: 22913716 Free PMC article.
-
Roles of c-type cytochromes in respiration in Neisseria meningitidis.Microbiology (Reading). 2008 Sep;154(Pt 9):2857-2864. doi: 10.1099/mic.0.2008/020339-0. Microbiology (Reading). 2008. PMID: 18757819
-
The unusual redox properties of C-type oxidases.Biochim Biophys Acta. 2016 Dec;1857(12):1892-1899. doi: 10.1016/j.bbabio.2016.09.009. Epub 2016 Sep 21. Biochim Biophys Acta. 2016. PMID: 27664317
-
Oxygen as Acceptor.EcoSal Plus. 2015;6(2):10.1128/ecosalplus.ESP-0012-2015. doi: 10.1128/ecosalplus.ESP-0012-2015. EcoSal Plus. 2015. PMID: 26734697 Free PMC article. Review.
-
Cytochrome cbb(3) oxidase and bacterial microaerobic metabolism.Biochem Soc Trans. 2002 Aug;30(4):653-8. doi: 10.1042/bst0300653. Biochem Soc Trans. 2002. PMID: 12196157 Review.
Cited by
-
Delta-proteobacterial SAR324 group in hydrothermal plumes on the South Mid-Atlantic Ridge.Sci Rep. 2016 Mar 8;6:22842. doi: 10.1038/srep22842. Sci Rep. 2016. PMID: 26953077 Free PMC article.
-
Molecular understanding of heteronuclear active sites in heme-copper oxidases, nitric oxide reductases, and sulfite reductases through biomimetic modelling.Chem Soc Rev. 2021 Mar 1;50(4):2486-2539. doi: 10.1039/d0cs01297a. Chem Soc Rev. 2021. PMID: 33475096 Free PMC article. Review.
-
The two transmembrane helices of CcoP are sufficient for assembly of the cbb3-type heme-copper oxygen reductase from Vibrio cholerae.Biochim Biophys Acta. 2015 Oct;1847(10):1231-9. doi: 10.1016/j.bbabio.2015.06.013. Epub 2015 Jun 25. Biochim Biophys Acta. 2015. PMID: 26116881 Free PMC article.
-
Cytochrome c4 is required for siderophore expression by Legionella pneumophila, whereas cytochromes c1 and c5 promote intracellular infection.Microbiology (Reading). 2011 Mar;157(Pt 3):868-878. doi: 10.1099/mic.0.046490-0. Epub 2010 Dec 22. Microbiology (Reading). 2011. PMID: 21178169 Free PMC article.
-
Structure of the bc1-cbb3 respiratory supercomplex from Pseudomonas aeruginosa.Proc Natl Acad Sci U S A. 2023 Oct 3;120(40):e2307093120. doi: 10.1073/pnas.2307093120. Epub 2023 Sep 26. Proc Natl Acad Sci U S A. 2023. PMID: 37751552 Free PMC article.
References
-
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. - PMC - PubMed
-
- Hemp J, Christian C, Barquera B, Gennis RB, Martinez TJ. Helix Switching of a Key Active-Site Residue in the Cytochrome cbb3 Oxidases. Biochemistry. 2005;44:10766–10775. - PubMed
-
- Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. - PubMed
-
- Hemp J, Han H, Roh JH, Kaplan S, Martinez TJ, Gennis RB. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 oxidase) of heme-copper oxygen reductases. Biochemistry. 2007;46:9963–9972. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

