Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion
- PMID: 20716517
- PMCID: PMC3001086
- DOI: 10.1093/nar/gkq720
Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion
Abstract
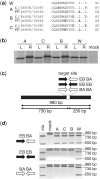
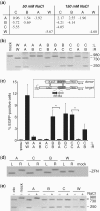
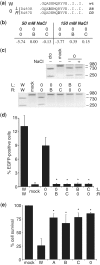
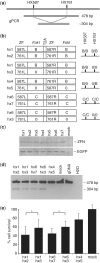
Zinc-finger nucleases (ZFNs) have been successfully used for rational genome engineering in a variety of cell types and organisms. ZFNs consist of a non-specific FokI endonuclease domain and a specific zinc-finger DNA-binding domain. Because the catalytic domain must dimerize to become active, two ZFN subunits are typically assembled at the cleavage site. The generation of obligate heterodimeric ZFNs was shown to significantly reduce ZFN-associated cytotoxicity in single-site genome editing strategies. To further expand the application range of ZFNs, we employed a combination of in silico protein modeling, in vitro cleavage assays, and in vivo recombination assays to identify autonomous ZFN pairs that lack cross-reactivity between each other. In the context of ZFNs designed to recognize two adjacent sites in the human HOXB13 locus, we demonstrate that two autonomous ZFN pairs can be directed simultaneously to two different sites to induce a chromosomal deletion in ∼ 10% of alleles. Notably, the autonomous ZFN pair induced a targeted chromosomal deletion with the same efficacy as previously published obligate heterodimeric ZFNs but with significantly less toxicity. These results demonstrate that autonomous ZFNs will prove useful in targeted genome engineering approaches wherever an application requires the expression of two distinct ZFN pairs.
Figures
Similar articles
-
Creating designed zinc-finger nucleases with minimal cytotoxicity.J Mol Biol. 2011 Jan 21;405(3):630-41. doi: 10.1016/j.jmb.2010.10.043. Epub 2010 Nov 19. J Mol Biol. 2011. PMID: 21094162 Free PMC article.
-
Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases.Nat Biotechnol. 2007 Jul;25(7):786-93. doi: 10.1038/nbt1317. Epub 2007 Jul 1. Nat Biotechnol. 2007. PMID: 17603476
-
Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells.Nucleic Acids Res. 2005 Oct 26;33(18):5978-90. doi: 10.1093/nar/gki912. Print 2005. Nucleic Acids Res. 2005. PMID: 16251401 Free PMC article. Review.
-
A novel zinc-finger nuclease platform with a sequence-specific cleavage module.Nucleic Acids Res. 2012 Mar;40(6):2623-38. doi: 10.1093/nar/gkr1112. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135304 Free PMC article.
-
[Zinc finger nucleases and their application].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010 Apr;27(2):162-5. doi: 10.3760/cma.j.issn.1003-9406.2010.02.010. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010. PMID: 20376797 Review. Chinese.
Cited by
-
Designed nucleases for targeted genome editing.Plant Biotechnol J. 2016 Feb;14(2):448-62. doi: 10.1111/pbi.12465. Epub 2015 Sep 15. Plant Biotechnol J. 2016. PMID: 26369767 Free PMC article. Review.
-
The application of genome editing in studying hearing loss.Hear Res. 2015 Sep;327:102-8. doi: 10.1016/j.heares.2015.04.016. Epub 2015 May 15. Hear Res. 2015. PMID: 25987504 Free PMC article. Review.
-
DNA cleavage enzymes for treatment of persistent viral infections: recent advances and the pathway forward.Virology. 2014 Apr;454-455:353-61. doi: 10.1016/j.virol.2013.12.037. Epub 2014 Jan 31. Virology. 2014. PMID: 24485787 Free PMC article. Review.
-
Points of View on the Tools for Genome/Gene Editing.Int J Mol Sci. 2021 Sep 13;22(18):9872. doi: 10.3390/ijms22189872. Int J Mol Sci. 2021. PMID: 34576035 Free PMC article. Review.
-
Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells.Nucleic Acids Res. 2015 Feb 18;43(3):e21. doi: 10.1093/nar/gku1246. Epub 2014 Nov 20. Nucleic Acids Res. 2015. PMID: 25414332 Free PMC article.
References
-
- Cathomen T, Schambach A. Zinc positive: tailored genome engineering meets reprogramming. Gene Ther. 2010;17:1–3. - PubMed
-
- Cathomen T, Joung KJ. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. - PubMed
-
- Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

