AUF1 p42 isoform selectively controls both steady-state and PGE2-induced FGF9 mRNA decay
- PMID: 20716519
- PMCID: PMC3001084
- DOI: 10.1093/nar/gkq717
AUF1 p42 isoform selectively controls both steady-state and PGE2-induced FGF9 mRNA decay
Abstract
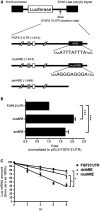
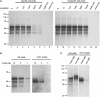
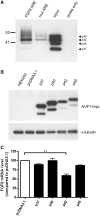
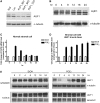
Fibroblast growth factor 9 (FGF9) is an autocrine/paracrine growth factor that plays vital roles in many physiologic processes including embryonic development. Aberrant expression of FGF9 causes human diseases and thus it highlights the importance of controlling FGF9 expression; however, the mechanism responsible for regulation of FGF9 expression is largely unknown. Here, we show the crucial role of an AU-rich element (ARE) in FGF9 3'-untranslated region (UTR) on controlling FGF9 expression. Our data demonstrated that AUF1 binds to this ARE to regulate FGF9 mRNA stability. Overexpression of each isoform of AUF1 (p37, p40, p42 and p45) showed that only the p42 isoform reduced the steady-state FGF9 mRNA. Also, knockdown of p42(AUF1) prolonged the half-life of FGF9 mRNA. The induction of FGF9 mRNA in prostaglandin (PG) E(2)-treated human endometrial stromal cells was accompanied with declined cytoplasmic AUF1. Nevertheless, ablation of AUF1 led to sustained elevation of FGF9 expression in these cells. Our study demonstrated that p42(AUF1) regulates both steady-state and PGE(2)-induced FGF9 mRNA stability through ARE-mediated mRNA degradation. Since almost half of the FGF family members are ARE-containing genes, our findings also suggest that ARE-mediated mRNA decay is a common pathway to control FGFs expression, and it represents a novel RNA regulon to coordinate FGFs homeostasis in various physiological conditions.
Figures


References
-
- Goldfarb M. Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev. 1996;7:311–325. - PubMed
-
- Gerwins P, Skoldenberg E, Claesson-Welsh L. Function of fibroblast growth factors and vascular endothelial growth factors and their receptors in angiogenesis. Crit. Rev. Oncol. Hematol. 2000;34:185–194. - PubMed
-
- Allouche M, Bikfalvi A. The role of fibroblast growth factor-2 (FGF-2) in hematopoiesis. Prog. Growth Factor Res. 1995;6:35–48. - PubMed
-
- Galzie Z, Kinsella AR, Smith JA. Fibroblast growth factors and their receptors. Biochem. Cell Biol. 1997;75:669–685. - PubMed

