Characterization of the Brain 26S Proteasome and its Interacting Proteins
- PMID: 20717473
- PMCID: PMC2901091
- DOI: 10.3389/fnmol.2010.00012
Characterization of the Brain 26S Proteasome and its Interacting Proteins
Abstract
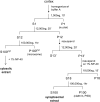
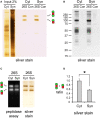
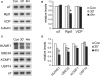
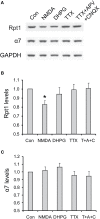
Proteasome-mediated proteolysis is important for synaptic plasticity, neuronal development, protein quality control, and many other processes in neurons. To define proteasome composition in brain, we affinity purified 26S proteasomes from cytosolic and synaptic compartments of the rat cortex. Using tandem mass spectrometry, we identified the standard 26S subunits and a set of 28 proteasome-interacting proteins that associated substoichiometrically and may serve as regulators or cofactors. This set differed from those in other tissues and we also found several proteins that associated only with either the cytosolic or the synaptic proteasome. The latter included the ubiquitin-binding factor TAX1BP1 and synaptic vesicle protein SNAP-25. Native gel electrophoresis revealed a higher proportion of doubly-capped 26S proteasome (19S-20S-19S) in the cortex than in the liver or kidney. To investigate the interplay between proteasome regulation and synaptic plasticity, we exposed cultured neurons to glutamate receptor agonist NMDA. Within 4 h, this agent caused a prolonged decrease in the activity of the ubiquitin-proteasome system as shown by disassembly of 26S proteasomes, decrease in ubiquitin-protein conjugates, and dissociation of the ubiquitin ligases UBE3A (E6-AP) and HUWE1 from the proteasome. Surprisingly, the regulatory 19S particles were rapidly degraded by proteasomal, not lysosomal degradation, and the dissociated E3 enzymes also degraded. Thus the content of proteasomes and their set of associated proteins can be altered by neuronal activity, in a manner likely to influence synaptic plasticity and learning.
Keywords: NMDA; UBE3A; proteasome; proteasome-interacting protein; synaptic plasticity; ubiquitin ligase.
Figures






Similar articles
-
Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes.Mol Biol Cell. 2000 Oct;11(10):3425-39. doi: 10.1091/mbc.11.10.3425. Mol Biol Cell. 2000. PMID: 11029046 Free PMC article.
-
[Comparative analysis of proteins associated with 26S and 20S proteasomes isolated from rabbit brain and liver].Biomed Khim. 2022 Jan;68(1):18-31. doi: 10.18097/PBMC20226801018. Biomed Khim. 2022. PMID: 35221293 Russian.
-
Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c.Proc Natl Acad Sci U S A. 2017 Apr 25;114(17):E3404-E3413. doi: 10.1073/pnas.1701734114. Epub 2017 Apr 10. Proc Natl Acad Sci U S A. 2017. PMID: 28396413 Free PMC article.
-
[Ubiquitin-independent protein degradation in proteasomes].Biomed Khim. 2018 Mar;64(2):134-148. doi: 10.18097/PBMC20186402134. Biomed Khim. 2018. PMID: 29723144 Review. Russian.
-
Proteasomes of the yeast S. cerevisiae: genes, structure and functions.Mol Biol Rep. 1995;21(1):3-10. doi: 10.1007/BF00990964. Mol Biol Rep. 1995. PMID: 7565661 Review.
Cited by
-
A Potential Mechanism for Targeting Aggregates With Proteasomes and Disaggregases in Liquid Droplets.Front Aging Neurosci. 2022 Apr 6;14:854380. doi: 10.3389/fnagi.2022.854380. eCollection 2022. Front Aging Neurosci. 2022. PMID: 35517053 Free PMC article. Review.
-
A high-throughput approach for measuring temporal changes in the interactome.Nat Methods. 2012 Sep;9(9):907-9. doi: 10.1038/nmeth.2131. Epub 2012 Aug 5. Nat Methods. 2012. PMID: 22863883 Free PMC article.
-
T2D and Depression Risk Gene Proteasome Modulator 9 is Linked to Insomnia.Sci Rep. 2015 Jul 13;5:12032. doi: 10.1038/srep12032. Sci Rep. 2015. PMID: 26166263 Free PMC article.
-
Network Analysis of UBE3A/E6AP-Associated Proteins Provides Connections to Several Distinct Cellular Processes.J Mol Biol. 2018 Mar 30;430(7):1024-1050. doi: 10.1016/j.jmb.2018.01.021. Epub 2018 Feb 6. J Mol Biol. 2018. PMID: 29426014 Free PMC article.
-
Detection of active proteasome structures in brain extracts: proteasome features of August rat brain with violations in monoamine metabolism.Oncotarget. 2017 Aug 10;8(41):70941-70957. doi: 10.18632/oncotarget.20208. eCollection 2017 Sep 19. Oncotarget. 2017. PMID: 29050334 Free PMC article.
References
-
- Bedford L., Hay D., Devoy A., Paine S., Powe D. G., Seth R., Gray T., Topham I., Fone K., Rezvani N., Mee M., Soane T., Layfield R., Sheppard P. W., Ebendal T., Usoskin D., Lowe J., Mayer R. J. (2008a). Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 28, 8189–819810.1523/JNEUROSCI.2218-08.2008 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

