Complex function by design using spatially pre-structured synthetic microbial communities: degradation of pentachlorophenol in the presence of Hg(ii)
- PMID: 20717565
- PMCID: PMC3005148
- DOI: 10.1039/c0ib00019a
Complex function by design using spatially pre-structured synthetic microbial communities: degradation of pentachlorophenol in the presence of Hg(ii)
Abstract
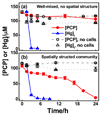
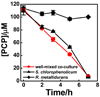
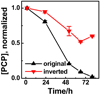
Naturally occurring microbes perform a variety of useful functions, with more complex processes requiring multiple functions performed by communities of multiple microbes. Synthetic biology via genetic engineering may be used to achieve desired multiple functions, e.g. multistep chemical and biological transformations, by adding genes to a single organism, but this is sometimes not possible due to incompatible metabolic requirements or not desirable in certain applications, especially in medical or environmental applications. Achieving multiple functions by mixing microbes that have not evolved to function together may not work due to competition of microbes, or lack of interactions among microbes. In nature, microbial communities are commonly spatially structured. Here, we tested whether spatial structure can be used to create a community of microbes that can perform a function they do not perform individually or when simply mixed. We constructed a core-shell fiber with Sphingobium chlorophenolicum, a pentachlorophenol (PCP) degrader, in the core layer and Ralstonia metallidurans, a mercuric ion (Hg(ii)) reducer, in the shell layer as a structured community using microfluidic laminar flow techniques. We applied a mixture of PCP and Hg(ii) to either a simple well-mixed culture broth (i.e. the unstructured community) or the spatially structured core-shell fibers. We found that without spatial structure, the community was unable to degrade PCP in the presence of Hg(ii) because S. chlorophenolicum is sensitive to Hg(ii). In contrast, with spatial structure in a core-shell fiber system, S. chlorophenolicum in a core layer was protected by R. metallidurans deposited in a shell layer, and the community was able to completely remove both PCP and Hg(ii) from a mixture. The appropriate size of the core-shell fiber was determined by the Damköhler number-the timescale of removal of Hg(ii) was on the same order of the timescale of diffusion of Hg(ii) through the outer layer when the shell layer was on the order of ~200 μm. Ultimately, with the ease of a child putting together 'Legos' to build a complex structure, using this approach one may be able to put together microorganisms to build communities that perform functions in vitro or even in vivo, e.g. as in a "microbiome on a pill".
Figures


Similar articles
-
Rhizoremediation of pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723.Chemosphere. 2007 Jun;68(5):864-70. doi: 10.1016/j.chemosphere.2007.02.014. Epub 2007 Mar 21. Chemosphere. 2007. PMID: 17376504
-
Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723.Appl Environ Microbiol. 2004 Apr;70(4):2391-7. doi: 10.1128/AEM.70.4.2391-2397.2004. Appl Environ Microbiol. 2004. PMID: 15066836 Free PMC article.
-
Biodegradation of hexachlorobenzene by a constructed microbial consortium.World J Microbiol Biotechnol. 2015 Feb;31(2):371-7. doi: 10.1007/s11274-014-1789-7. Epub 2014 Dec 23. World J Microbiol Biotechnol. 2015. PMID: 25532745
-
Bacterial resistances to inorganic mercury salts and organomercurials.Plasmid. 1992 Jan;27(1):4-16. doi: 10.1016/0147-619x(92)90002-r. Plasmid. 1992. PMID: 1311113 Review.
-
Molecular analysis of pentachlorophenol degradation.Biodegradation. 1994 Dec;5(3-4):277-88. doi: 10.1007/BF00696465. Biodegradation. 1994. PMID: 7765838 Review.
Cited by
-
Engineered Living Hydrogels.Adv Mater. 2022 Jul;34(26):e2201326. doi: 10.1002/adma.202201326. Epub 2022 Apr 24. Adv Mater. 2022. PMID: 35243704 Free PMC article. Review.
-
Emerging strategies for engineering microbial communities.Biotechnol Adv. 2019 Nov 1;37(6):107372. doi: 10.1016/j.biotechadv.2019.03.011. Epub 2019 Mar 15. Biotechnol Adv. 2019. PMID: 30880142 Free PMC article. Review.
-
Engineering microbial systems to explore ecological and evolutionary dynamics.Curr Opin Biotechnol. 2012 Oct;23(5):791-7. doi: 10.1016/j.copbio.2012.01.006. Epub 2012 Feb 4. Curr Opin Biotechnol. 2012. PMID: 22310174 Free PMC article. Review.
-
Deciphering microbial spatial organization: insights from synthetic and engineered communities.ISME Commun. 2025 Jun 27;5(1):ycaf107. doi: 10.1093/ismeco/ycaf107. eCollection 2025 Jan. ISME Commun. 2025. PMID: 40821451 Free PMC article. Review.
-
Modelling compartmentalization towards elucidation and engineering of spatial organization in biochemical pathways.Sci Rep. 2017 Sep 21;7(1):12057. doi: 10.1038/s41598-017-11081-8. Sci Rep. 2017. PMID: 28935941 Free PMC article.
References
-
- Brenner K, You LC, Arnold FH. Trends Biotechnol. 2008;26:483–489. - PubMed
-
- Hasty J, McMillen D, Collins JJ. Nature. 2002;420:224–230. - PubMed
-
- Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Curr. Opin. Biotechnol. 2008;19:556–563. - PubMed
-
- de Lorenzo V. Curr. Opin. Biotechnol. 2008;19:579–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

