Ralstonia solanacearum ΔPGI-1 strain KZR-5 is affected in growth, response to cold stress and invasion of tomato
- PMID: 20717661
- PMCID: PMC3011089
- DOI: 10.1007/s00248-010-9728-0
Ralstonia solanacearum ΔPGI-1 strain KZR-5 is affected in growth, response to cold stress and invasion of tomato
Abstract
The survival and persistence of Ralstonia solanacearum biovar 2 in temperate climates is still poorly understood. To assess whether genomic variants of the organism show adaptation to local conditions, we compared the behaviour of environmental strain KZR-5, which underwent a deletion of the 17.6 kb genomic island PGI-1, with that of environmental strain KZR-1 and potato-derived strains 1609 and 715. PGI-1 harbours two genes of potential ecological relevance, i.e. one encoding a hypothetical protein with a RelA/SpoT domain and one a putative cellobiohydrolase. We thus assessed bacterial fate under conditions of amino acid starvation, during growth, upon incubation at low temperature and invasion of tomato plants. In contrast to the other strains, environmental strain KZR-5 did not grow on media that induce amino acid starvation. In addition, its maximum growth rate at 28°C in rich medium was significantly reduced. On the other hand, long-term survival at 4°C was significantly enhanced as compared to that of strains 1609, 715 and KZR-1. Although strain KZR-5 showed growth rates (at 28°C) in two different media, which were similar to those of strains 1609 and 715, its ability to compete with these strains under these conditions was reduced. In singly inoculated tomato plants, no significant differences in invasiveness were observed among strains KZR-5, KZR-1, 1609 and 715. However, reduced competitiveness of strain KZR-5 was found in experiments on tomato plant colonisation and wilting when using 1:1 or 5:1 mixtures of strains. The potential role of PGI-1 in plant invasion, response to stress and growth in competition at high and moderate temperatures is discussed.
Figures

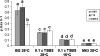
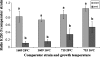
Similar articles
-
A putative genomic island, PGI-1, in Ralstonia solanacearum biovar 2 revealed by subtractive hybridization.Antonie Van Leeuwenhoek. 2010 Oct;98(3):359-77. doi: 10.1007/s10482-010-9450-4. Epub 2010 May 14. Antonie Van Leeuwenhoek. 2010. PMID: 20467813 Free PMC article.
-
The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato.mBio. 2012 Aug 31;3(4):e00114-12. doi: 10.1128/mBio.00114-12. Print 2012. mBio. 2012. PMID: 22807564 Free PMC article.
-
Ralstonia solanacearum requires PopS, an ancient AvrE-family effector, for virulence and To overcome salicylic acid-mediated defenses during tomato pathogenesis.mBio. 2013 Nov 26;4(6):e00875-13. doi: 10.1128/mBio.00875-13. mBio. 2013. PMID: 24281716 Free PMC article.
-
Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence.BMC Genomics. 2010 Jun 15;11:379. doi: 10.1186/1471-2164-11-379. BMC Genomics. 2010. PMID: 20550686 Free PMC article.
-
Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen-limited host xylem environment.mBio. 2015 Mar 17;6(2):e02471. doi: 10.1128/mBio.02471-14. mBio. 2015. PMID: 25784703 Free PMC article.
Cited by
-
Proteomic comparison of Ralstonia solanacearum strains reveals temperature dependent virulence factors.BMC Genomics. 2014 Apr 12;15:280. doi: 10.1186/1471-2164-15-280. BMC Genomics. 2014. PMID: 24725348 Free PMC article.
-
Complete Genome Sequence of Sequevar 14M Ralstonia solanacearum Strain HA4-1 Reveals Novel Type III Effectors Acquired Through Horizontal Gene Transfer.Front Microbiol. 2019 Aug 14;10:1893. doi: 10.3389/fmicb.2019.01893. eCollection 2019. Front Microbiol. 2019. PMID: 31474968 Free PMC article.
-
Ralstonia solanacearum Facing Spread-Determining Climatic Temperatures, Sustained Starvation, and Naturally Induced Resuscitation of Viable but Non-Culturable Cells in Environmental Water.Microorganisms. 2022 Dec 16;10(12):2503. doi: 10.3390/microorganisms10122503. Microorganisms. 2022. PMID: 36557756 Free PMC article.
References
-
- Calderón-Flores A, Du Pont G, Huerta-Saquero A, Merchant-Larios H, Servin-Gonzalez L, Duran S. The stringent response is required for amino acid and nitrate utilization, nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J Bacteriol. 2005;187:5075–5083. doi: 10.1128/JB.187.15.5075-5083.2005. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

