Coverage statistics for sequence census methods
- PMID: 20718980
- PMCID: PMC2940910
- DOI: 10.1186/1471-2105-11-430
Coverage statistics for sequence census methods
Abstract
Background: We study the statistical properties of fragment coverage in genome sequencing experiments. In an extension of the classic Lander-Waterman model, we consider the effect of the length distribution of fragments. We also introduce a coding of the shape of the coverage depth function as a tree and explain how this can be used to detect regions with anomalous coverage. This modeling perspective is especially germane to current high-throughput sequencing experiments, where both sample preparation protocols and sequencing technology particulars can affect fragment length distributions.
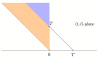
Results: Under the mild assumptions that fragment start sites are Poisson distributed and successive fragment lengths are independent and identically distributed, we observe that, regardless of fragment length distribution, the fragments produced in a sequencing experiment can be viewed as resulting from a two-dimensional spatial Poisson process. We then study the successive jumps of the coverage function, and show that they can be encoded as a random tree that is approximately a Galton-Watson tree with generation-dependent geometric offspring distributions whose parameters can be computed.
Conclusions: We extend standard analyses of shotgun sequencing that focus on coverage statistics at individual sites, and provide a null model for detecting deviations from random coverage in high-throughput sequence census based experiments. Our approach leads to explicit determinations of the null distributions of certain test statistics, while for others it greatly simplifies the approximation of their null distributions by simulation. Our focus on fragments also leads to a new approach to visualizing sequencing data that is of independent interest.
Figures



References
-
- Weber J, Myers E. Human whole-genome shotgun sequencing. Genome Research. 1997;7:401–409. - PubMed
-
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, Anson EL, Bolanos RA, Chou HH, Jordan CM, Halpern AL, Lonardi S, Beasley EM, Brandon RC, Chen L, Dunn PJ, Lai Z, Liang Y, Nusskern DR, Zhan M, Zhang Q, Zheng X, Rubin GM, Adams MD, Venter JC. A Whole-Genome Assembly of Drosophila. Science. 2000;287(5461):2196–2204. doi: 10.1126/science.287.5461.2196. - DOI - PubMed

