Epstein-Barr virus nuclear antigen 1 Hijacks the host kinase CK2 to disrupt PML nuclear bodies
- PMID: 20719947
- PMCID: PMC2953151
- DOI: 10.1128/JVI.01183-10
Epstein-Barr virus nuclear antigen 1 Hijacks the host kinase CK2 to disrupt PML nuclear bodies
Abstract
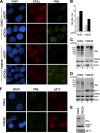
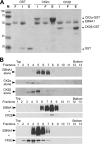
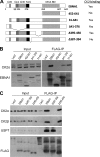
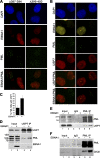
Latent Epstein-Barr virus (EBV) infection is an important causative factor in the development of several cancers, including nasopharyngeal carcinoma (NPC). The one EBV protein expressed in the nucleus of NPC cells, EBNA1, has been shown to disrupt promyelocitic leukemia (PML) nuclear bodies (NBs) by inducing the degradation of PML proteins, leading to impaired DNA repair and increased cell survival. Although EBNA1-mediated PML disruption is likely to be an important factor in the development of NPC, little is known about its mechanism. We now show that an interaction between EBNA1 and the host CK2 kinase is crucial for EBNA1 to disrupt PML bodies and degrade PML proteins. EBNA1 increases the association of CK2 with PML proteins, thereby increasing the phosphorylation of PML proteins by CK2, a modification that is known to trigger the polyubiquitylation and degradation of PML. The interaction between EBNA1 and CK2 is direct and occurs through the β regulatory subunit of CK2 and EBNA1 amino acids 387 to 394. The binding of EBNA1 to the host ubiquitin specific protease USP7 has also been shown to be important for EBNA1-mediated PML disruption. We show that EBNA1 also increases the occupancy of USP7 at PML NBs and that CK2 and USP7 bind independently and simultaneously to EBNA1 to form a ternary complex. The combined results indicate that EBNA1 usurps two independent cellular pathways to trigger the loss of PML NBs.
Figures


References
-
- Appel, K., P. Wagner, B. Boldyreff, O. G. Issinger, and M. Montenarh. 1995. Mapping of the interaction sites of the growth suppressor protein p53 with the regulatory beta-subunit of protein kinase CK2. Oncogene 11:1971-1978. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

