A conserved domain in the coronavirus membrane protein tail is important for virus assembly
- PMID: 20719948
- PMCID: PMC2953170
- DOI: 10.1128/JVI.01131-10
A conserved domain in the coronavirus membrane protein tail is important for virus assembly
Abstract
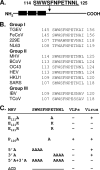
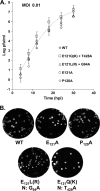
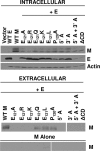
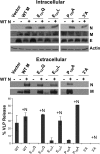
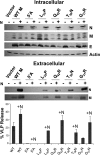
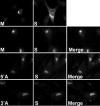
Coronavirus membrane (M) proteins play key roles in virus assembly, through M-M, M-spike (S), and M-nucleocapsid (N) protein interactions. The M carboxy-terminal endodomain contains a conserved domain (CD) following the third transmembrane (TM) domain. The importance of the CD (SWWSFNPETNNL) in mouse hepatitis virus was investigated with a panel of mutant proteins, using genetic analysis and transient-expression assays. A charge reversal for negatively charged E(121) was not tolerated. Lysine (K) and arginine (R) substitutions were replaced in recovered viruses by neutrally charged glutamine (Q) and leucine (L), respectively, after only one passage. E121Q and E121L M proteins were capable of forming virus-like particles (VLPs) when coexpressed with E, whereas E121R and E121K proteins were not. Alanine substitutions for the first four or the last four residues resulted in viruses with significantly crippled phenotypes and proteins that failed to assemble VLPs or to be rescued into the envelope. All recovered viruses with alanine substitutions in place of SWWS residues had second-site, partially compensating, changes in the first TM of M. Alanine substitution for proline had little impact on the virus. N protein coexpression with some M mutants increased VLP production. The results overall suggest that the CD is important for formation of the viral envelope by helping mediate fundamental M-M interactions and that the presence of the N protein may help stabilize M complexes during virus assembly.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

