Apoptosis of hepatitis B virus-infected hepatocytes prevents release of infectious virus
- PMID: 20719950
- PMCID: PMC2977891
- DOI: 10.1128/JVI.00653-10
Apoptosis of hepatitis B virus-infected hepatocytes prevents release of infectious virus
Abstract
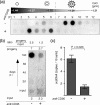
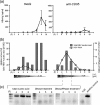
Apoptosis of infected cells is critically involved in antiviral defense. Apoptosis, however, may also support the release and spread of viruses. Although the elimination of infected hepatocytes is required to combat hepatitis B virus (HBV) infection, it is still unknown which consequences hepatocyte apoptosis has for the virus and whether or not it is advantageous to the virus. To study this, we designed a cell culture model consisting of both HBV-producing cell lines and primary human hepatocytes serving as an infection model. We showed that the release of mature, enveloped virions was 80% to 90% reduced 24 h after the induction of apoptosis in HBV-replicating hepatoma cells or HBV-infected hepatocytes. Importantly, HBV particles released from apoptotic hepatocytes were immature and nonenveloped and proved not to be infectious. We found an inverse correlation between the strength of an apoptotic stimulus and the infectivity of the virus particles released: the more potent the apoptotic stimulus, the higher the ratio of nonenveloped capsids to virions and the lower their infectivity. Furthermore, we demonstrated that HBV replication and, particularly, the expression of the HBx protein transcribed from the viral genome during replication do not sensitize cells to apoptosis. Our data clearly reject the hypothesis that the apoptosis of infected hepatocytes facilitates the propagation of HBV. Rather, these data indicate that HBV needs to prevent the apoptosis of its host hepatocyte to ensure the release of infectious progeny and, thus, virus spread in the liver.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

