13C-tracer and gas chromatography-mass spectrometry analyses reveal metabolic flux distribution in the oleaginous microalga Chlorella protothecoides
- PMID: 20720172
- PMCID: PMC2948989
- DOI: 10.1104/pp.110.158956
13C-tracer and gas chromatography-mass spectrometry analyses reveal metabolic flux distribution in the oleaginous microalga Chlorella protothecoides
Abstract
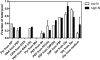
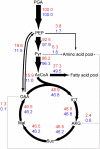
The green alga Chlorella protothecoides has received considerable attention because it accumulates neutral triacylglycerols, commonly regarded as an ideal feedstock for biodiesel production. In order to gain a better understanding of its metabolism, tracer experiments with [U-(13)C]/[1-(13)C]glucose were performed with heterotrophic growth of C. protothecoides for identifying the metabolic network topology and estimating intracellular fluxes. Gas chromatography-mass spectrometry analysis tracked the labeling patterns of protein-bound amino acids, revealing a metabolic network consisting of the glycolysis, the pentose phosphate pathway, and the tricarboxylic acid cycle with inactive glyoxylate shunt. Evidence of phosphoenolpyruvate carboxylase, phosphoenolpyruvate carboxykinase, and malic enzyme activity was also obtained. It was demonstrated that the relative activity of the pentose phosphate pathway to glycolysis under nitrogen-limited environment increased, reflecting excess NADPH requirements for lipid biosynthesis. Although the growth rate and cellular oil content were significantly altered in response to nitrogen limitation, global flux distribution of C. protothecoides remained stable, exhibiting the rigidity of central carbon metabolism. In conclusion, quantitative knowledge on the metabolic flux distribution of oleaginous alga obtained in this study may be of value in designing strategies for metabolic engineering of desirable bioproducts.
Figures
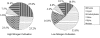
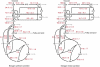
References
-
- Benthin S, Nielsen J, Villadsen J. (1991) A simple and reliable method for the determination of cellular RNA content. Biotechnol Tech 5: 39–42
-
- Botham PA, Ratledge C. (1979) Biochemical explanation for lipid accumulation in Candida-107 and other oleaginous microorganisms. J Gen Microbiol 114: 361–375 - PubMed
-
- Brune DE, Lundquist TJ, Benemann JR. (2009) Microalgal biomass for greenhouse gas reductions: potential for replacement of fossil fuels and animal feeds. J Environ Eng 135: 1136–1144
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

