Tumor necrosis factor-α represses the expression of NHE2 through NF-κB activation in intestinal epithelial cell model, C2BBe1
- PMID: 20722069
- PMCID: PMC2990806
- DOI: 10.1002/ibd.21419
Tumor necrosis factor-α represses the expression of NHE2 through NF-κB activation in intestinal epithelial cell model, C2BBe1
Abstract
Background: High levels of proinflammatory cytokines are linked to pathogenesis of diarrhea in inflammatory bowel disease (IBD). Na(+) absorption is compromised in IBD. The studies were designed to determine the effect of tumor necrosis factor-α (TNF-α) on the expression and activity of NHE2, a Na(+) /H(+) exchanger (NHE) that is involved in transepithelial Na(+) absorption in intestinal epithelial cells.
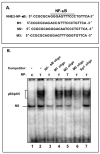
Methods: NHE2 regulation was examined in TNF-α-treated C2BBe1 cells by reverse-transcription polymerase chain reaction (RT-PCR), reporter gene assays, and Western blot analysis. NHE isoform activities were measured as ethyl-isopropyl-amiloride- and HOE694-sensitive (22) Na-uptake. In vitro and in vivo protein-DNA interactions were assessed by gel mobility shift assays and chromatin immunoprecipitation studies.
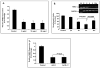
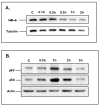
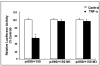
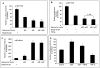
Results: TNF-α treatment of C2BBe1 cells led to repression of NHE2 promoter activity, mRNA, and protein levels; and inhibited both NHE2 and NHE3 mediated (22) Na-uptake. 5'-deletion analysis of the NHE2 promoter-reporter constructs identified basepair -621 to -471 as the TNF-α-responsive region (TNF-RE). TNF-α activated NF-κB subunits, p50 and p65, and their DNA-binding to a putative NF-κB motif within TNF-RE. Mutations in the NF-κB motif abolished NF-κB-DNA interactions and abrogated TNF-α-induced repression. Ectopic overexpression of NF-κB resulted in repression of NHE2 expression. Two functionally distinct inhibitors of NF-κB blocked the inhibitory effect of TNF-α.
Conclusions: The human NHE2 isoform is a direct target of transcription factor NF-κB. TNF-α-mediated activation of NF-κB decreases the expression and activity of NHE2 in the intestinal epithelial cell line, C2BBe1. These findings implicate NF-κB in the modulation of Na(+) absorption during intestinal inflammatory conditions such as IBD where a high level of TNF-α is detected.
Copyright © 2010 Crohn's & Crohn's & Colitis Foundation of America, Inc.
Figures

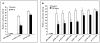


References
-
- Zachos NC, Tse M, Donowitz M. Molecular physiology of intestinal Na+/H+ exchange. Annu Rev Physiol. 2005;67:411–43. - PubMed
-
- Malakooti J, Dahdal RY, Dudeja PK, et al. The human Na(+)/H(+) exchanger NHE2 gene: genomic organization and promoter characterization. Am J Physiol Gastrointest Liver Physiol. 2001;280:G763–73. - PubMed
-
- Malakooti J, Sandoval R, Memark VC, et al. Zinc finger transcription factor Egr-1 is involved in stimulation of NHE2 gene expression by phorbol 12-myristate 13-acetate. Am J Physiol Gastrointest Liver Physiol. 2005;289:G653–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
