Using text to build semantic networks for pharmacogenomics
- PMID: 20723615
- PMCID: PMC2991587
- DOI: 10.1016/j.jbi.2010.08.005
Using text to build semantic networks for pharmacogenomics
Abstract
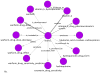
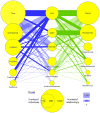
Most pharmacogenomics knowledge is contained in the text of published studies, and is thus not available for automated computation. Natural Language Processing (NLP) techniques for extracting relationships in specific domains often rely on hand-built rules and domain-specific ontologies to achieve good performance. In a new and evolving field such as pharmacogenomics (PGx), rules and ontologies may not be available. Recent progress in syntactic NLP parsing in the context of a large corpus of pharmacogenomics text provides new opportunities for automated relationship extraction. We describe an ontology of PGx relationships built starting from a lexicon of key pharmacogenomic entities and a syntactic parse of more than 87 million sentences from 17 million MEDLINE abstracts. We used the syntactic structure of PGx statements to systematically extract commonly occurring relationships and to map them to a common schema. Our extracted relationships have a 70-87.7% precision and involve not only key PGx entities such as genes, drugs, and phenotypes (e.g., VKORC1, warfarin, clotting disorder), but also critical entities that are frequently modified by these key entities (e.g., VKORC1 polymorphism, warfarin response, clotting disorder treatment). The result of our analysis is a network of 40,000 relationships between more than 200 entity types with clear semantics. This network is used to guide the curation of PGx knowledge and provide a computable resource for knowledge discovery.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

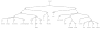
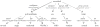
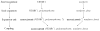
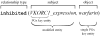



Similar articles
-
PGxCorpus, a manually annotated corpus for pharmacogenomics.Sci Data. 2020 Jan 2;7(1):3. doi: 10.1038/s41597-019-0342-9. Sci Data. 2020. PMID: 31896797 Free PMC article.
-
A knowledge-driven conditional approach to extract pharmacogenomics specific drug-gene relationships from free text.J Biomed Inform. 2012 Oct;45(5):827-34. doi: 10.1016/j.jbi.2012.04.011. Epub 2012 Apr 27. J Biomed Inform. 2012. PMID: 22561026 Free PMC article.
-
Inferring the semantic relationships of words within an ontology using random indexing: applications to pharmacogenomics.AMIA Annu Symp Proc. 2013 Nov 16;2013:1123-32. eCollection 2013. AMIA Annu Symp Proc. 2013. PMID: 24551397 Free PMC article.
-
Essential Characteristics of Pharmacogenomics Study Publications.Clin Pharmacol Ther. 2019 Jan;105(1):86-91. doi: 10.1002/cpt.1279. Clin Pharmacol Ther. 2019. PMID: 30406943 Free PMC article. Review.
-
Systematic review of the evidence on the cost-effectiveness of pharmacogenomics-guided treatment for cardiovascular diseases.Genet Med. 2020 Mar;22(3):475-486. doi: 10.1038/s41436-019-0667-y. Epub 2019 Oct 8. Genet Med. 2020. PMID: 31591509 Free PMC article.
Cited by
-
Trends in computational biology—2010.Nat Biotechnol. 2011 Jan;29(1):45-9. doi: 10.1038/nbt.1747. Nat Biotechnol. 2011. PMID: 21221103 No abstract available.
-
Representing annotation compositionality and provenance for the Semantic Web.J Biomed Semantics. 2013 Nov 22;4:38. doi: 10.1186/2041-1480-4-38. eCollection 2013. J Biomed Semantics. 2013. PMID: 24268021 Free PMC article.
-
Automatic construction of a large-scale and accurate drug-side-effect association knowledge base from biomedical literature.J Biomed Inform. 2014 Oct;51:191-9. doi: 10.1016/j.jbi.2014.05.013. Epub 2014 Jun 10. J Biomed Inform. 2014. PMID: 24928448 Free PMC article.
-
A mutation-centric approach to identifying pharmacogenomic relations in text.J Biomed Inform. 2012 Oct;45(5):835-41. doi: 10.1016/j.jbi.2012.05.003. Epub 2012 Jun 7. J Biomed Inform. 2012. PMID: 22683993 Free PMC article.
-
Learning the Structure of Biomedical Relationships from Unstructured Text.PLoS Comput Biol. 2015 Jul 28;11(7):e1004216. doi: 10.1371/journal.pcbi.1004216. eCollection 2015 Jul. PLoS Comput Biol. 2015. PMID: 26219079 Free PMC article.
References
-
- Klein T, Chang J, Cho M, Easton K, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver D, Rubin D, Shafa F, Stuart J, Altman R. Integrating genotype and phenotype information: An overview of the PharmGKB project. The Pharmacogenomics Journal. 1:167–170. - PubMed
-
- Blaschke C, Andrade MA, Ouzounis C, Valencia A. Automatic Extraction of Biological Information from Scientific Text: Protein–Protein Interactions. ISMB; 1999. pp. 60–67. - PubMed
-
- Rosario B, Hearst MA. Classifying semantic relations in bioscience texts. ACL; 2004. pp. 430–437.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

