ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death
- PMID: 20723759
- PMCID: PMC2925044
- DOI: 10.1016/j.cell.2010.07.018
ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death
Abstract
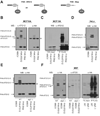
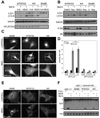
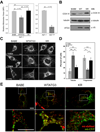
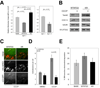
ATG12, an ubiquitin-like modifier required for macroautophagy, has a single known conjugation target, another autophagy regulator called ATG5. Here, we identify ATG3 as a substrate for ATG12 conjugation. ATG3 is the E2-like enzyme necessary for ATG8/LC3 lipidation during autophagy. ATG12-ATG3 complex formation requires ATG7 as the E1 enzyme and ATG3 autocatalytic activity as the E2, resulting in the covalent linkage of ATG12 onto a single lysine on ATG3. Surprisingly, disrupting ATG12 conjugation to ATG3 does not affect starvation-induced autophagy. Rather, the lack of ATG12-ATG3 complex formation produces an expansion in mitochondrial mass and inhibits cell death mediated by mitochondrial pathways. Overall, these results unveil a role for ATG12-ATG3 in mitochondrial homeostasis and implicate the ATG12 conjugation system in cellular functions distinct from the early steps of autophagosome formation.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol. 2009 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

